Abstract
Metastatic melanoma (MM) displays remarkable phenotypic plasticity, allowing tumor cells to transition reversibly between proliferative and mesenchymal (MES)-like states. This dynamic switching is strongly associated with therapeutic resistance and poor prognosis. Although transcriptional and epigenetic mechanisms driving these transitions have been extensively studied, the role of post-translational regulation, particularly the ubiquitin–proteasome system, remains poorly understood. Here, we identify the ubiquitin E3 ligase RNF 123 (KPC1) as a key post-translational suppressor of MES reprogramming in MM. Integrative analyses of bulk and single-cell transcriptomic datasets revealed that KPC1 expression is inversely correlated with the expression of core mesenchymal markers such as ZEB1, CDH2, and AXL, and positively associated with epithelial and melanocytic lineage genes, including CDH1 and MITF. Deconvolution of TCGA-SKCM RNA-seq data confirmed that this inverse correlation is specific to malignant melanoma cells and strongest in tumors enriched for mesenchymal gene signatures. Single-cell trajectory and enrichment analyses further demonstrated that decreasing KPC1 expression accompanies MES-like switch. Mechanistically, KPC1 binds and promotes the ubiquitination and proteasomal-mediated degradation of ZEB1, thereby suppressing cadherin switching and cell motility. Loss of KPC1 in melanoma cells prevented ZEB1 proteasomal-mediated degradation, increased expression of mesenchymal markers, and enhanced MM cells migration. Clinically, low KPC1 protein levels were associated with increased expression of ZEB1 and CDH2 and poorer overall survival. Furthermore, combined assessment of KPC1, ZEB1, and CDH2 expression improved patient stratification, suggesting the potential utility of multi-marker signatures for prognostic modeling. These findings establish KPC1 as a central post-translational regulator of melanoma cell state plasticity through targeted degradation of ZEB1. This study highlights a novel mechanism regulating MES-like transition and highlights KPC1 as a potential theragnostic target in MM.

Similar content being viewed by others
Introduction
Cutaneous melanoma is a highly aggressive malignancy, particularly in patients with metastatic disease [1]. Melanoma incidence has been steadily increasing in Australia, Western Europe, and the United States [1,2,3]. The current first-line systemic treatment for metastatic melanoma (MM) involves immune checkpoint inhibitors (ICI) and/or targeted therapies, and more recently cell-based therapies [4,5,6]. The prognosis for patients with melanoma tumors that has metastasized to distant sites, such as visceral organs or the brain, remains dismal, with a 5-years survival rate ~35% [7, 8]. A central biological feature underlying this aggressiveness is the phenotypic plasticity of melanoma cells which implies their capacity to reversibly switch among a melanocytic, a proliferative phenotype and an invasive, and a mesenchymal (MES)-like states [9]. These cell states transition shares transcriptional features with the epithelial-to-mesenchymal transition (EMT), including loss of differentiation markers and upregulation of mesenchymal drivers, and has been linked to enhanced motility, therapeutic resistance, and metastatic potential [10, 11]. The MES-like phenotypic state is typically marked by downregulation of the microphthalmia-associated transcription factor (MITF) and upregulation of markers such as AXL receptor tyrosine kinase (AXL) and zinc finger e-box binding homeobox1 (ZEB1) [12, 13]. However, despite detailed transcriptomic characterization, the upstream mechanisms that govern this reversible switch remains poorly defined.
Although the phenotypic plasticity of melanoma has been established, the molecular circuits regulating transitions between proliferative and MES-like states are still being elucidated [14]. Signaling cascades such as the MAPK, TGF-β, and Wnt pathways have been implicated in promoting MES features and invasiveness in melanoma cells [15,16,17], yet these do not fully explain the dynamic reversibility or intra-tumoral heterogeneity observed in melanoma progression. ZEB1, a key transcriptional regulator of EMT in carcinomas, also plays a prominent role in the MES state of melanoma, repressing genes and facilitating an invasive behavior [18]. However, the regulatory mechanisms controlling ZEB1 expression and protein turnover in this context are poorly defined. Recent work has revealed that melanoma cell state is governed by multi-modal regulatory programs, integrating chromatin remodeling, metabolic status, transcription factor networks, and immune signaling, all of which may influence transitions between distinct cellular phenotypes [19]. Within this framework, non-genetic mechanisms —including microRNA-mediated suppression and proteasomal degradation via ubiquitin ligases— have also emerged as important contributors to phenotype switching [9, 20, 21]. Still, the involvement of the ubiquitin–proteasome system in modulating MES associated gene expression patterns in MM remains underexplored.
Kip1 ubiquitylation-promoting complex subunit 1 (KPC1), also known as RNF123, is an E3 ubiquitin ligase that targets the NF-κB precursor protein p105 for proteasomal processing, thereby modulating inflammatory and survival pathways [22, 23]. Beyond its canonical function in NF-κB regulation, KPC1 has been implicated in diverse tumor-suppressive roles across multiple cancer types, including glioblastoma, breast, and prostate cancer, by controlling cell cycle regulators and affecting EMT components [24, 25]. For instance, it has been shown to mediate ubiquitination of p27 Kip1 and Vimentin, suggesting its involvement in cell cycle control and inhibition of mesenchymal programs [26, 27]. However, no prior studies have examined whether KPC1 modulates the MES transcriptional program or phenotype switching in MM.
In this study, we investigated the role of KPC1 in regulating phenotypic plasticity in MM. Through integrative analysis combining bulk and single-cell transcriptomics, multiplex immunofluorescence (mIF), and functional assays, we demonstrated that KPC1 negatively regulates the MES program by promoting the ubiquitin-dependent degradation of ZEB1 —a central driver of mesenchymal reprogramming. Downregulation of KPC1 results in ZEB1 stabilization, increased expression of MES markers, and enhanced cellular migration. Downregulation of KPC1 is also associated with poor clinical outcomes in MM patients. Our findings position KPC1 as a gatekeeper of melanoma cell state transitions and suggest that restoration or mimicking of KPC1 activity may represent a novel therapeutic approach to suppress melanoma progression and metastasis.
Materials and methods
Melanoma cell lines
Established MM cell lines from SJCI were attained from melanoma patients who received elective surgery (DP-0574, FD-0836, HM-0525, LP-0024, ML-0817, MH-0331, M-12, M204, VN-0326, WP-0614). The cell lines were cultured in RPMI-1640 and supplemented with 10 mM HEPES, 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin (complete medium). All human cell lines have been authenticated using short tandem repeat (STR) profiling within the last three years. All experiments were performed with mycoplasma-free cell lines.
Public datasets
Bulk RNA sequencing (RNA-seq) datasets from The Cancer Genome Atlas (TCGA) [28] and Genotype-Tissue Expression (GTEx) [29] were downloaded through the website of University of California Santa Cruz (UCSC) Xena [30]. Single-cell RNA-seq (scRNA-seq) dataset of melanoma was obtained from GSE115978 [31], which includes transcriptomic profiles of 31 melanoma tumors from patients with metastatic melanoma who were treated with immune checkpoint inhibitors (ICI). The protein expression dataset of melanoma patients was obtained from PXD006003 [32], which includes proteomic profiles of advanced-stage melanoma patients undergoing tumor-infiltrating lymphocyte (TIL)-based or anti-PD1 immunotherapy, quantified using high-resolution mass spectrometry.
Deconvolution analysis of the bulk RNA sequencing data sets
BayesPrism [33] and CODEFACS [34] deconvolution analyses were applied to the bulk RNA sequencing (RNA-seq) data set of TCGA-SKCM [30] to obtain melanoma cell-specific expression profiles. For BayesPrism, the count matrixes of messenger RNA (mRNA) expression profiles were provided as input and the raw count matrix of the single cell RNA-seq (scRNA-seq) data set from GSE115978 [31] was used as the reference gene expression profile. For CODEFACS, the mRNA expression profiles of Transcripts Per Million (TPM) format were provided as input and the same gene expression profile generated in the previous study was used as reference [34]. CODEFACS deconvolution analysis was used to get the immune cell fractions. For CODEFACS, the Leukocyte Signature Matrix 22 (LM22) signature matrix [35] was used as the reference gene expression profiles of immune cells. LM22 was downloaded from the CIBERSORTx web portal (https://cibersortx.stanford.edu/) [35].
Pseudospatial reconstruction of single-cell transcriptomes
Pseudospatial reconstruction of single-cell transcriptomes was performed using Monocle3 algorithm [36, 37]. Cells were ordered based on genes identified through differential gene expression analysis, comparing inner and outer cell fractions. Significant genes were selected using a likelihood ratio test, with criteria of FDR < 1 × 10⁻¹⁰ and |log₂ fold-change | > 1. These genes were used as “ordering genes” to construct trajectories with Monocle3 functions (setOrderingFilter(), reduceDimension(), and orderCells()). MES marker expression across pseudospace was visualized using the plot_genes_in_pseudotime function [36, 37].
Statistical analysis
Variations among variables were compared using either the Mann–Whitney U test, Student’s t-test, or Fisher’s exact test where applicable. Multiple groups were analyzed by one-way ANOVA followed by Tukey’s post hoc test. The Kaplan–Meier method was utilized to estimate overall survival (OS), progression-free survival (PFS) and disease-specific survival (DSS), and comparisons between survival curves were made using the Log-rank test. The statistical analyses were two-sided and performed using R software v4.4.2. A significance level of p < 0.05 was considered statistically significant. All experiments were performed in triplicate unless otherwise noted.
Additional M&M and uncropped western blot images can be found in Supplementary Information, Table S1 and S2, Supplementary Material files. All Western blot quantifications are provided in Figs. S7 and S8.
Results
KPC1 expression is inversely associated with key mesenchymal markers in MM
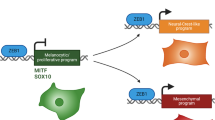
To examine the relationship between KPC1 expression and mesenchymal reprogramming in MM, we employed a five-pronged integrative strategy (Fig. 1A). As an initial step, we performed a comprehensive correlation analysis between KPC1 mRNA levels and mesenchymal gene signatures across 33 cancer types, including samples from the TCGA-SKCM cohort. Although correlation patterns varied among tumor types, KPC1 mRNA levels generally exhibited a negative association with MES marker genes (Figs. 1B and S1A, B). Building on a previous classification of MM into melanocytic (MEL), neural/plastic (NPLAS), MES clusters based on MES-associated gene signatures [19], we next compared KPC1 expression across these transcriptomic-based molecular subtypes. Notably, KPC1 expression was significantly reduced in the MES cluster (Fig. 1C, D). In contrast, ZEB1, CDH2, and AXL mRNA levels —canonical MES markers— showed the highest levels in the MES cluster (Fig. 1E, F; Fig. S2A, C), whereas CDH1 and MITF expression were the lowest in MES cluster (Fig. S2B, D). We further stratified MM into KPC1-high and KPC1-low groups. In the low-KPC1 group, ZEB1, AXL, and CDH2 were significantly upregulated (Fig. S2E, F, H), while CDH1 and MITF were significantly upregulated in the high-KPC1 group (Fig. S2G, I). Correlation analyses confirmed that KPC1 was significantly inversed correlated with ZEB1, AXL, and CDH2 (Fig. S2J, K, M), and positively correlated with CDH1 and MITF (Fig. S2L, N).
A Schematic overview of multimodal approaches used to define mesenchymal (MES) state switching in metastatic melanoma (MM). B Left: Heatmap of Spearman correlation between KPC1 and 149 MES signature genes across 33 TCGA tumor types; a red arrow highlights TCGA-SKCM dataset. Right: Dot plot of Spearman’s correlation values (R) between KPC1 and MES signature genes in each tumor type; circle size indicates the fraction of significant genes negative or positive correlated with KPC1. C Box plot of KPC1 mRNA (log2) in TCGA-SKCM samples stratified as MEL, NPLAS, or MES. D Box plot of KPC1 mRNA (log2) in TCGA-SKCM samples stratified as MES or non-MES. E ZEB1 mRNA (log2) in MEL, NPLAS, and MES subsets of TCGA-SKCM dataset. F ZEB1 expression in non-MES versus MES subsets of TCGA-SKCM dataset. Data are presented as the mean ± standard deviation. Three-group comparisons: one-way ANOVA with Tukey’s post hoc test.
To validate these findings at the protein level, we analyzed data from the PXD006003 proteomic dataset. Consistent with the transcriptomic observations, KPC1 protein levels were inversely correlated with MES markers (Fig. S1C, D). Together, these findings support a robust inverse relationship between KPC1 expression and MES gene programs in MM at both the mRNA and protein levels.
Deconvolution analysis reveals a negative correlation between KPC1 and genes in melanoma cells
Bulk RNA-seq data contains a mixture of the expression profiles from various cell types, making it potentially insufficient to clearly determine that the relationship between KPC1 expression and MES marker genes occur specifically in tumor cells. To address this limitation, deconvolution analysis was performed on MM samples from the TCGA-SKCM RNA-seq dataset using CODEFACS and BayesPrism pipelines. The deconvolution analysis with CODEFACS identified ten distinct cell types (Fig. 2A). The highest KPC1 mRNA levels were observed in the malignant melanoma cell fraction (Fig. 2A). Among the MEL, NPLAS, and MES clusters, KPC1 expression was the lowest in the MES cluster (Fig. 2B). In contrast, ZEB1 and CDH2 exhibited the highest expression levels in the MES cluster, whereas CDH1 expression was also reduced (Fig. 2B). Similarly, BayesPrism-based deconvolution analysis also confirmed that malignant melanoma cells exhibited the highest KPC1 expression levels (Fig. 2C). Among the three clusters, BayesPrism-based analysis yielded consistent results, showing that KPC1 expression was lowest in the MES cluster (Fig. 2D). Conversely, ZEB1 and CDH2 displayed the highest expression levels in the MES cluster, while CDH1 expression was also lower in this group (Fig. 2D). Subsequently, single-sample gene set enrichment analysis (ssGSEA) was conducted using the deconvoluted melanoma cell profiles from MM samples obtained from the TCGA-SKCM dataset to explore signaling pathways associated with KPC1. These analyses revealed that KPC1 levels in MM cells were negatively correlated with the ssGSEA hallmark EMT pathway score (Fig. 2E, F). In summary, these findings indicate that KPC1 expression in MM cells is negatively correlated with MES marker genes, suggesting a link between KPC1 levels and the mesenchymal status in MM cells.
A Spearman’s correlation between infiltrated immune cell fraction levels and KPC1 mRNA levels in metastatic melanoma cells, estimated by CODEFACS. B Box plot of KPC1, ZEB1, CDH2, and CDH1 mRNA levels in MEL, NPLAS, and MES clusters, estimated by CODEFACS. C Distribution of KPC1 mRNA levels in different cell types, estimated by BayesPrism. D Box plot of KPC1, ZEB1, CDH2, and CDH1 mRNA levels in MEL, NPLAS, and MES clusters, estimated by BayesPrism. E Spearman’s correlation between KPC1 mRNA levels and pathway enrichment scores, calculated by ssGSEA in TCGA-SKCM metastatic melanoma samples. F Spearman’s correlation coefficient (R) between KPC1 mRNA levels and EMT pathway enrichment scores, calculated by ssGSEA in TCGA-SKCM metastatic melanoma samples. Three-group comparisons: one-way ANOVA with Tukey’s post hoc test.
Single-cell RNA-seq analysis highlights the association between KPC1 and MES marker genes in MM
To further investigate the association between KPC1 and mesenchymal marker genes in MM cells, we performed a detailed analysis using scRNA-seq data from MM. scRNA-seq analysis identified nine distinct cell types (Fig. 3A), where malignant melanoma cells showed the highest KPC1 expression (Fig. 3B). Additionally, pathway analysis revealed that MES related pathways were significantly enriched in the KPC1 low-expression group (Fig. 3C). To explore the relationship between KPC1 and MES in greater detail, trajectory analysis was conducted using the scRNA-seq data. Importantly, there was an MES enrichment followed by a decrease in KPC1 expression (Fig. 3D). Furthermore, MES-promoting factors such as ZEB1, CDH2, and VIM were upregulated with MES enrichment, whereas MES-suppressing factors like CDH1 were downregulated as MES became more enriched (Fig. 3E). In summary, these findings provide strong evidence that KPC1 expression is inversely associated with MES enrichment in MM. This is accompanied by the upregulation of MES-promoting genes and the downregulation of MES-suppressing genes, suggesting a dynamic interplay between KPC1 expression and the transition to a MES-like state in MM cells.
A t-SNE plot showing the clustering of nine distinct cell types based on single-cell RNA-seq data from metastatic melanoma (GSE115978). Colors represent post hoc annotation of single-cell profiles. B Distribution of KPC1 mRNA levels across cell types. Left: Heatmap showing KPC1 expression in different cell types. Right: Dot plots illustrate KPC1 mRNA levels in melanoma cells compared to other cell types. Single-cell expression profiles were re-clustered using Seurat (v4.0.5). C Pathway enrichment analysis of melanoma cells with low KPC1 expression group. Left: Hallmark enrichment pathway. Right: REACTOME enrichment pathway. D Left: Trajectory analysis was conducted using single-cell RNA-seq data to examine the relationship between KPC1 expression and MES enrichment. Right: The analysis illustrates the progression of MES and the corresponding changes in KPC1 expression. Statistical significance of the trajectory was evaluated using Moran’s I. E Expression patterns of mesenchymal marker genes, including MES-promoting factors (ZEB1, CDH2, and VIM) and MES-suppressing factors (CDH1), were evaluated along the trajectory of MES enrichment. Statistical significance of the trajectory was evaluated using Moran’s I.
KPC1 silencing drives post-transcriptional stabilization of ZEB1 and MES reprogramming
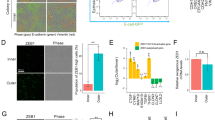
To investigate the functional consequences of KPC1 downregulation, we examined MM cell lines previously classified into MEL, NPLAS, and MES transcriptomic subtypes [19] (Fig. 4A). Our focus was on determining whether p65 or KPC1-mediated regulation of p50 levels had any effect on ZEB1 transcriptional regulation. Silencing of canonical NF-κB pathway component p65 did not affect ZEB1 expression at either the mRNA or protein level (Fig. S3A–D). KPC1-knockdown led to robust depletion of KPC1, accumulation of its canonical substrates p105, and a decrease in p50 (Fig. 4B), consistent with our previous reports [25], but ZEB1 mRNA levels remained unchanged (Fig. 4C, D), confirming that NF-κB signaling is dispensable for ZEB1 transcriptional regulation. In contrast, baseline KPC1 protein inversely correlated with ZEB1 abundance across ten MM cell lines (Fig. 4E, F). KPC1- knockdown induced a dose-dependent stabilization of ZEB1 and CDH2, along with reduced CDH1 expression, indicative of a cadherin switch (Figs. 4G–I and S3E, F). Functionally, KPC1-deficient cells showed enhanced migratory activity in wound-healing assays (Fig. 4J, K). In a previous study, VIM was reported to be a substrate for KPC1 [27]; however, VIM protein levels were unchanged in MM cell lines with KPC1 downregulation (Fig. S3H). MITF is a key transcriptional factor associated with a melanocytic lineage program [19]. Based on the MES-like phenotype switching, we evaluated MITF levels to determine whether KPC1 knockdown in MM cell lines. Consistently with a MES-like phenotype switching, MITF decreased at both the mRNA and protein levels (Fig. S3G, H). These data support the role for KPC1 in suppressing MES reprogramming via post-transcriptional regulation of ZEB1 and maintaining MITF levels.
A Heatmap showing relative expression (z-score) of 89 melanocytic-like (MEL) and 113 mesenchymal-like (MES) signature genes across ten metastatic melanoma (MM) cell lines, clustered into MEL, neural/plastic (NPLAS), and MES subtypes. B Western blot images show KPC1, p105, and p50 protein levels following siRNA-mediated silencing of KPC1 (siKPC1) in HM-0525 and DP-0574 cell lines compared to control conditions (siCtrl). C, D qRT-PCR analysis of KPC1 and ZEB1 in HM-0525 (C) and DP-0574 (D) cells transfected with siCtrl or siKPC1. E Western blot images show KPC1 and ZEB1 protein levels in various MM cell lines. F Scatter plot showing the correlation levels between endogenous KPC1 and ZEB1 protein levels (relative to β-actin) across the ten MM cell lines; subtypes colored as in (A), Pearson’s correlation coefficient (r) and p-value indicated. G, H Western blot images of KPC1, ZEB1, CDH2, and CDH1 following siKPC1 at varying concentrations (1, 5, 10, 25 nM) in HM-0525 and DP-0574 cell lines. I Western blot images of KPC1, ZEB1, CDH2, and CDH1 protein levels following siKPC1 vs siCtrl in HM-0525 and DP-0574 cell lines. J, K Wound healing assays using HM-0525 and DP-0527 cells transfected with siCtrl or siKPC1. The migrated distance was quantified by measuring the difference at 0, 24, 48, and 72 h and was normalized to 0-hour timepoint. Data are presented as the mean ± standard deviation. qRT-PCR data were analyzed by two-tailed unpaired Student’s t-test. Wound healing data were analyzed by two-way repeated-measures ANOVA with Bonferroni post-hoc correction for multiple comparisons. These data represent three independent experiments, each conducted in triplicate. N.S. not significant.
Next, we examined whether externally TGF-β induced MES-like switching changes the KPC1 abundance in MM cell lines. The TGF-β induced MES-like state in MM cell lines was characterized by a spindle morphology, ZEB1/CDH2 upregulation and CDH1 downregulation, together with a decrease in KPC1 protein (Fig. S4A, B). These results suggest that TGF-β induced MES-like switching may be a potential trigger of KPC1 downregulation, which consequently initiates a MES-like phenotype switching in MM.
KPC1 mediates ubiquitin-dependent proteasome turnover of ZEB1 in MM cells
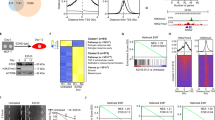
Based on the ZEB1 protein regulatory effect mediated by KPC1 in MM cell lines, we next assessed whether KPC1 controls ZEB1 protein turnover. Cycloheximide (CHX) chase assays showed that KPC1- knockdown delayed ZEB1 degradation over 6 h, indicating that KPC1 loss stabilizes ZEB1 (Fig. 5A, B). Proteasome inhibition using MG132 further confirmed that ZEB1 is degraded via the ubiquitin–proteasome system. MG132 treatment increased ZEB1 levels and induced accumulation of polyubiquitinated ZEB1 species (Fig. 5C, D). Co-immunoprecipitation revealed that endogenous KPC1 interacted with endogenous ZEB1 in MM cells (Fig. 5E). In immunoprecipitation assay using ubiquitin-trap beads, KPC1- knockdown was linked to a reduced level of ubiquitinated-ZEB1 (Fig. 5F). Together, these findings demonstrated that KPC1 promotes the ubiquitin-dependent proteasomal degradation of ZEB1 in MM cells and suggested that KPC1 may represent an E3-ligase mediating ZEB1 ubiquitination.
A, B Cycloheximide (CHX) chase in HM-0525 (A) and DP-0574 (B) cells transfected with control siRNA (siCtrl) or KPC1 siRNA (siKPC1). Cells were treated with CHX (50 µg/mL) for 0, 2, 4, or 6 h. Upper panels show representative Western blots images for KPC1 and ZEB1; β-actin is loading control. Lower panels plot mean ± SD of ZEB1/β-actin from three independent experiments. C, D Proteasome inhibition in CHX-treated cells. HM-0525 (C) and DP-0574 (D) cells were cotreated with CHX (50 µg/mL) and MG132 (10 µM) for 0, 1, 2, or 4 h. Left, Western blots images for ZEB1 (top), ubiquitin (Ub; middle), and β-actin (bottom). Right, quantification of ZEB1/β-actin (mean ± SD, n = 3); statistical comparisons versus 0 h by one-way ANOVA with Dunnett’s post hoc test. E Immunoprecipitation of endogenous KPC1. HM-0525 and DP-0574 cells were treated with MG132 (10 µM, 4 h), lysed, and subjected to IP with anti-KPC1 or control IgG. Whole-cell lysate (WCL), IgG IP, and KPC1 IP fractions were Western-blotted for KPC1 (inset) and ZEB1; β-actin is loading control. Heavy-chain and light-chain bands of the antibody used in IP are indicated. F Ubiquitin-Trap assay for ZEB1 ubiquitination. DP-0574 and HM-0525 cells transfected with siCtrl or siKPC1 were treated with MG132 (10 µM, 4 h). Ubiquitin-binding beads captured ubiquitinated proteins from cleared lysates; eluates and WCL were analyzed by Western blot using for total Ubiquitin (top), ZEB1 (middle) and KPC1 (bottom) protein levels; β-actin served as loading control.
Then, we wondered whether ZEB1 transcription factor controls KPC1 protein levels in a feedback loop. To address that MM cell lines with high endogenous levels of ZEB1 and low endogenous levels of KPC1 were treated with siRNA targeting ZEB1. Surprisingly, ZEB1 downregulation did not affect the protein levels of KPC1 (Fig. S4C–E). These results suggest that KPC1 protein levels are not affected by ZEB1 downregulation to maintain the MES-like phenotypes in MM cell lines.
KPC1 loss correlates with MES marker upregulation and poor prognosis in MM
To assess the relationship between KPC1 downregulation and MES marker expression in tumors from MM patients, a TMA was stained for MART1, KPC1, ZEB1 and CDH2 by mIF. 110 cores from MM patients were evaluated for staining after quality control. MM tumors with high KPC1 protein showed robust KPC1 signals that were accompanied by low levels of ZEB1 and CDH2; conversely, MM tumors with low KPC1 exhibited low KPC1 protein levels, but markedly increased ZEB1 and CDH2 protein levels (Fig. 6A). Quantitative image analysis across all cores revealed a significant inverse correlation between KPC1 and ZEB1 protein levels (Fig. 6B), and ZEB1 abundance was higher in the KPC1-low group compared to KPC1-high group (Fig. 6C). We next evaluated the prognostic impact of KPC1, ZEB1 and CDH2 protein expression using the clinical annotations for the MM patients included in the TMA. MM patients with low KPC1 expression experienced markedly poorer overall survival (OS, Fig. 6D). Likewise, high ZEB1 or high CDH2 protein levels in the tumor cells (MART-1+) were associated with a worse prognosis (Fig. 6E, F). MM patients having low KPC1 expression and high ZEB1 protein levels exhibited a significantly poorer prognosis (Fig. 6G). Together, these data demonstrated that KPC1 downregulation in MM correlates with MES marker upregulation. KPC1, ZEB1, and CDH2 carry independent prognostic value in MM patients.
A Representative multiplex immunofluorescence images from TMA cores stratified by KPC1 protein expression (“High” vs. “Low”; n = 3 cores per group). Rows show single-channel DAPI, MART1 (cyan), KPC1 (green), ZEB1 (magenta), CDH2 (yellow), and the merged four-color overlay. Scale bars, 100 µm. B Scatter plot of KPC1 versus ZEB1 protein levels (n = 110 patients). Spearman’s correlation coefficient (R) and two-tailed p-value are indicated. C Box plots comparing ZEB1 protein expression between Low KPC1 (n = 55) and High KPC1 (n = 55) patient groups. D–G Kaplan–Meier analyses for overall survival (OS) when considering the protein levels of (D) KPC1, (E) ZEB1, (F) CDH2, and (G) combined KPC1/ZEB1 expression subgroups in 110 MM patients. Numbers in parentheses denote group sizes. Log-rank test p-values are indicated.
Role of KPC1 as a biomarker in SKCM and UVM
As previously described, KPC1 mRNA levels are downregulated in MM tissue [25]. In this exploratory analysis across solid tumors using TCGA dataset, we analyzed data from 33 tumor types and 30 corresponding normal tissues datasets. We demonstrated that KPC1 is highly expressed in tumor tissues compared to normal tissues across various cancer types (Fig. S5A). Additionally, patients were stratified based on KPC1 expression in multiple cancers, including MM. Of clinical relevance, low KPC1 expression was associated with significant worse OS, disease-specific survival (DSS), and progression-free survival (PFS), in both SKCM and UVM (Fig. S5B–D). These findings suggested that KPC1 has the potential to serve as a biomarker context dependent on specific cancer types. To determine whether expression of mesenchymal markers stratifies patient outcome in MM, we analyzed OS using TCGA-SKCM dataset according to ZEB1, CDH2 and CDH1 mRNA levels. Patients with high ZEB1 had significantly shorter OS than those with low ZEB1 (Fig. S6A). Similarly, high CDH2 expression predicted worse OS compared with low CDH2 (Fig. S6A), and low CDH1 was associated with inferior OS relative to high CDH1 (Fig. S6A). We next examined combined signatures: patients with low KPC1 and high ZEB1 fared markedly worse than those with high KPC1 and low ZEB1 (Fig. S6B), and inclusion of CDH2 further refined risk stratification, with the low KPC1/high ZEB1/high CDH2 group showing the poorer OS (Fig. S6B). These data demonstrate that elevated ZEB1 and CDH2, as well as reduced CDH1, portend worse clinical outcomes in MM, and that combined assessment of KPC1, ZEB1 and CDH2 expression can further improve prognostic precision.
Discussion
The present study identifies KPC1 as a key negative regulator of MES reprogramming in MM. Through integrative transcriptomic, proteomic, and functional analyses, we demonstrated that KPC1 expression is inversely associated with mesenchymal markers such as ZEB1 and CDH2, and AXL, and positively correlated with epithelial regulators including CDH1 and MITF. Mechanistically, KPC1 promotes the ubiquitination and proteasomal degradation of ZEB1, thereby limiting cadherin switching, cell migration, and the activation of a mesenchymal transcriptional program. These findings establish KPC1 as a post-translational gatekeeper of MES-like cell state in MM.
The results of this study extends the current mechanistic models of transcriptional and epigenetic control of melanoma phenotypic switching [38, 39], to demonstrate that ubiquitin–proteasome–mediated regulation of ZEB1 protein stability represents a critical and underexplored layer of control. This mechanism explains how rapid and reversible phenotypic transitions may occur, potentially independently of immediate alterations in mRNA levels. Intriguingly, while KPC1 is known to modulate NF-κB signaling via p105 processing to p50 [22], our findings indicate that KPC1-mediated suppression of ZEB1, a key driver of the MES state, occurs independently of the canonical NF-κB p65/p50 pathway. This is evidenced by the lack of ZEB1 alteration upon p65 silencing, suggesting a distinct, direct regulatory role for KPC1 in controlling melanoma plasticity via ZEB1 that is separate from its impact on NF-κB signaling. Furthermore, our findings suggest that KPC1 activity is intrinsically linked to the established MEL (MITF-high/AXL-low) versus MES (MITF-low/AXL-high) lineage states. Specifically, the higher KPC1 expression in tumors with melanocytic signatures and lower expression in MES-enriched tumors positions KPC1 as a potential upstream regulator or a key stabilizing factor for the melanocytic phenotype, possibly by suppressing ZEB1-driven mesenchymal programs that antagonize MITF expression or function. Conversely, a decrease in KPC1 levels may lower the threshold for cells to switch towards a ZEB1-high/MITF-low mesenchymal state, a transition strongly associated with increased invasiveness and therapy resistance. In our system, KPC1 loss was accompanied by reduced MITF at both transcript and protein levels while vimentin remained unchanged, supporting a lineage-shift interpretation rather than vimentin-mediated effects. Consistent with this view, exogenous induction of a MES-like program was associated with lower KPC1 in MM cell lines, suggesting that phenotype-switching cues can modulate KPC1 and thereby favor ZEB1 stabilization. In line with this framework, our prior work showed that upregulation of miR-155 reduces KPC1 in MM cells [25], and TGF-β has been reported to increase miR-155 and promote cell invasion [40], providing a plausible miR-155–mediated link between MES-like induction and reduced KPC1. Collectively, these results complement the existing models based on the dynamic interplay of known melanoma lineage-defining transcription factors like MITF, SOX10, BRN2, and AXL [10, 14], by adding KPC1 as a crucial checkpoint. Furthermore, our findings that KPC1 acts upstream to control ZEB1 stability align with and provide a novel regulatory mechanism for previous studies highlighting ZEB1 as a central mediator of invasive and therapy-resistant melanoma states [18].
Previous study by our group demonstrated that KPC1 is downregulated as melanoma patients progressed to advance stages [25]. As noted above, the downregulation of KPC1 could be explained by an upregulation of miR-155-5p in MM tumors that target KPC1 mRNA levels [25]. From a translational perspective, restoring or mimicking KPC1 function may offer a novel means to restrict tumor progression given the association between mesenchymal states and drug resistance, immune evasion, and metastasis [11, 14]. Therefore, potential strategies aimed at pharmacologic enhancement of KPC1 expression or ligase activity, as well as targeted degradation of ZEB1 via PROTACs or other modalities, may hold therapeutic value. Additionally, the combined expression profile of KPC1 and ZEB1 may serve as a prognostic biomarker, enabling improved risk stratification in patients with MM.
While our study provides robust and reproducible findings regarding the KPC1-ZEB1 axis using siRNA-mediated knockdown in in vitro models, we acknowledge certain limitations inherent to this approach. The use of cell lines, though informative for mechanistic dissection, may not fully recapitulate the complex tumor microenvironment and heterogeneity observed in vivo. Furthermore, while siRNA offers effective target downregulation, future studies employing alternative genetic manipulation techniques could further solidify the specific role of KPC1. Therefore, subsequent investigations will be valuable to ascertain the broader physiological and pathological relevance of KPC1-mediated regulation of tumor plasticity and its implications for therapeutic responses in more complex, clinically relevant settings.
In summary, this study uncovers that KPC1 is a suppressor of MES status in MM by mediating the ZEB1 ubiquitination and proteasomal degradation. These findings contribute to a growing understanding of post-translational regulation of the ZEB1 master regulator of MES status and tumor plasticity associated with resistance and immune evasion. KPC1 could be a potential target for theragnostic target development in MM patients undergoing targeted or ICIs therapies. The MES status of melanoma may be indicative of potential non-responsiveness to certain types of therapeutic interventions thus may serve as a triaging factor to specific therapies.
Data availability
Data are available in a public, open access repository. All datasets and databases utilized throught the manuscript are described in Table S1.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34.
Guy GP Jr, Thomas CC, Thompson T, Watson M, Massetti GM, Richardson LC. Vital signs: melanoma incidence and mortality trends and projections - United States, 1982-2030. MMWR Morb Mortal Wkly Rep. 2015;64:591–6.
Arnold M, Singh D, Laversanne M, Vignat J, Vaccarella S, Meheus F, et al. Global burden of cutaneous melanoma in 2020 and projections to 2040. JAMA Dermatol. 2022;158:495–503.
Gershenwald JE, Scolyer RA. Melanoma staging: american joint committee on cancer (AJCC) 8th edition and beyond. Ann Surg Oncol. 2018;25:2105–10.
Scolyer RA, Rawson RV, Gershenwald JE, Ferguson PM, Prieto VG. Melanoma pathology reporting and staging. Mod Pathol. 2020;33:15–24.
Seth R, Agarwala SS, Messersmith H, Alluri KC, Ascierto PA, Atkins MB, et al. Systemic therapy for melanoma: ASCO guideline update. J Clin Oncol. 2023;41:4794–820.
Gershenwald JE, Scolyer RA, Hess KR, Sondak VK, Long GV, Ross MI, et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67:472–92.
Weiss SA, Wolchok JD, Sznol M. Immunotherapy of melanoma: facts and hopes. Clin Cancer Res. 2019;25:5191–201.
Kozar I, Bai Y, Smith CA, Hoser M, Karki P, Wang J, et al. A UBR5–ZEB1–epithelial–mesenchymal transition axis in melanoma regulates phenotype switching and therapeutic resistance. Cell Rep. 2022;40:111256.
Tsoi J, Robert L, Paraiso K, Galvan C, Sheu KM, Lay J, et al. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell. 2018;33:890–904.
Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–6.
Verfaillie A, Imrichova H, Atak ZK, Dewaele M, Rambow F, Hulselmans G, et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Cell. 2015;161:1681–96.
Hoek KS, Eichhoff OM, Schlegel NC, Döbbeling U, Kobert N, Schaerer L, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–6.
Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M, et al. Toward minimal residual disease-directed therapy in melanoma. Nature. 2018;562:191–7.
Dissanayake SK, Olkhanud PB, O’Connell MP, Carter A, French AD, Camilli TC, et al. Wnt5A regulates expression of tumor-associated antigens in melanoma via changes in STAT3 phosphorylation. Oncogene. 2007;26:4780–9.
Zhuang L, Lee CS, Scolyer RA, McCarthy SW, Palmer AA, Zhang X-D, et al. Activation of the ERK pathway by TGF-β in human melanoma cells: Implications for progression and therapy. Oncogene. 2012;21:8194–207.
Anastas JN, Kulikauskas RM, Tamir T, Rizos H, Long GV, von Euw EM, et al. WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors. Cell Rep. 2014;124:66–79.
Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–80.
Andrews MC, Oba J, Wu C-J, Zhu H, Karpinets T, Creasy CA, et al. Multi-modal molecular programs regulate melanoma cell state. Nat Commun. 2022;13:4000.
Laurette P, Strub T, Koludrovic D, Keime C, Le Gras S, Seberg H, et al. Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. Nat Commun. 2015;6:6683.
Perotti V, Baldassari P, Molla A, Vegetti C, Bersani I, Anichini A. An actionable axis linking NFATc2 to EZH2 controls the EMT-like program of melanoma cells. Nat Med. 2019;25:1877–87.
Kravtsova-Ivantsiv Y, Shomer I, Cohen-Kaplan V, Snijder B, Superti-Furga G, Gonen H, et al. KPC1-Mediated Ubiquitination and Proteasomal Processing of NF-κB1 p105 to p50 Restricts Tumor Growth. Cell. 2015;161:333–47.
Wang X, Bustos MA, Zhang X, Ramos RI, Tan C, Iida Y, et al. Downregulation of the Ubiquitin-E3 Ligase RNF123 Promotes Upregulation of the NF-κB1 Target SerpinE1 in Aggressive Glioblastoma Tumors. Cancers (Basel) 2020;12.
Gulei D, Drula R, Ghiaur G, Buzoianu AD, Kravtsova-Ivantsiv Y, Tomuleasa C, et al. The tumor suppressor functions of ubiquitin ligase KPC1: from cell-cycle control to NF-κB Regulator. Cancer Res. 2023;83:1762–7.
Iida Y, Ciechanover A, Marzese DM, Hata K, Bustos M, Ono S, et al. Epigenetic regulation of KPC1 ubiquitin ligase affects the NF-κB pathway in melanoma. Clin Cancer Res. 2017;23:4831–42.
Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, et al. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol. 2004;6:1229–35.
Gottfried Y, Lulu-Shimron C, Goldhirsh G, Fisher Y, Ziv T, Hoon DSB, et al. Vimentin is a ubiquitination and degradation substrate of the ubiquitin ligase KPC1. Biochem Biophys Res Commun. 2024;745:151231.
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–20.
The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580-5.
Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8.
Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175:984–997.e924.
Harel M, Ortenberg R, Varanasi SK, Mangalhara KC, Mardamshina M, Markovits E, et al. Proteomics of Melanoma Response to Immunotherapy Reveals Mitochondrial Dependence. Cell. 2019;179:236–250.e218.
Chu T, Wang Z, Pe’er D, Danko CG. Cell type and gene expression deconvolution with BayesPrism enables Bayesian integrative analysis across bulk and single-cell RNA sequencing in oncology. Nat Cancer. 2022;3:505–17.
Wang K, Patkar S, Lee JS, Gertz EM, Robinson W, Schischlik F, et al. Deconvolving Clinically Relevant Cellular Immune Cross-talk from Bulk Gene Expression Using CODEFACS and LIRICS Stratifies Patients with Melanoma to Anti-PD-1 Therapy. Cancer Discov. 2022;12:1088–105.
Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–82.
Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502.
McFaline-Figueroa JL, Hill AJ, Qiu X, Jackson D, Shendure J, Trapnell C. A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition. Nat Genet. 2019;51:1389–98.
Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84.
Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45.
Li DP, Fan J, Wu YJ, Xie YF, Zha JM, Zhou XM. MiR-155 up-regulated by TGF-β promotes epithelial-mesenchymal transition, invasion and metastasis of human hepatocellular carcinoma cells in vitro. Am J Transl Res. 2017;9:2956–65.
Acknowledgements
The authors thank all staff of the Department of Translational Molecular Medicine at SJHC for their kind advisory and technical assistance.
Funding
DSBH and AC were supported by a collaborative grant from the Adelson Medical Research Foundation (AMRF). AC was supported also by grants from the Israel Science Foundation (ISF), from the Israel Personal Medicine Partnership (IPMP) administered by the ISF, from the Rappaport Foundation, and by a generous gift donated by Craig Darian and the late Albert Sweet and administered by the American Technion Society (ATS). A.C. is an Israel Cancer Research Fund (ICRF) USA Professor.
Author information
Authors and Affiliations
Contributions
Y Nakano: experimental design, data acquisition, data analysis, data interpretation, original manuscript drafting. MA Bustos: experimental design, data interpretation, scientific discussion, supervision, manuscript reviewing and editing. KK Chong: data acquisition, data analysis, scientific discussion, manuscript reviewing and editing. Y Hayashi: experimental design, data interpretation, scientific discussion. A Ciechanover: conceptualization, data interpretation, scientific discussion, supervision. DSB Hoon: conceptualization, data interpretation, scientific discussion, supervision, funding acquisition, manuscript reviewing and editing. All authors critically revised the article for important intellectual content and approved the submission for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study involves human participants and was approved by Ethics Committees: SJHC/SJCI Joint Institutional Review Board (IRB) and Western IRB. Approval ID#: MORD-RTPCR-0995. All methods were performed in accordance with the relevant guidelines and regulations. Participants gave informed consent to participate in the study before taking part.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Dr. Gerry Melino
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nakano, Y., Bustos, M.A., Chong, K.K. et al. Ubiquitin E3 ligase KPC1 governs mesenchymal metastatic melanoma reprogramming via proteasomal degradation of ZEB1. Cell Death Dis 16, 897 (2025). https://doi.org/10.1038/s41419-025-08262-z
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41419-025-08262-z