Abstract
Recent progress in cancer metabolism research has identified lactylation as a critical post-translational modification influencing tumor development and progression. The process relies on lactate accumulation and the activation of lactate-sensitive acyltransferases. Beyond its role in epigenetic regulation, lactylation has emerged as a significant factor in tumor metabolism and evolution, offering fresh opportunities for developing targeted therapies that transcend traditional approaches. This review explores the growing importance of lactylation in cancer biology and highlights its potential for advancing diagnostic tools and therapeutic strategies.
Similar content being viewed by others
FACTS
-
Lactylation, a novel post-translational modification utilizing lactate as a substrate, is crucial in regulating tumor metabolism, epigenetics, and the tumor immune microenvironment.
-
The accumulation of lactate is essential for the activation of lactate-sensing acyltransferases and the occurrence of lactylation. This metabolic preference of tumors serves as the cradle for lactylation, giving new significance to the Warburg effect.
-
Lactylation modifications span the entire course of tumor evolution, extending beyond regulating the tumor’s epigenetic modifications. They broadly affect tumor initiation, progression, and the remodeling of the tumor microenvironment.
-
Building on the foundation of previous research on metabolism and targeted drugs, leveraging lactate sensors and lactylation regulatory mechanisms as potential targets for cancer treatment offers distinct advantages.
Open questions
-
Q1 Acyltransferases Preference
For non-specific acyltransferases, is there a competitive relationship between acetyl-CoA and lactyl-CoA, and which acyltransferases prefer to bind?
-
Q2 Lactylation Erasers
If there are specific lactylation writers, are there specific lactylation erasers?
-
Q3 Order of Lactylation
In the tumor microenvironment, which occurs earlier: lactylation in immune cells or in the tumor cells themselves?
-
Q4 Role of Lactylation in Cancer
Does lactylation in tumors only have a promoting effect on cancer, is it a perfect target?
Introduction
The Warburg effect, a hallmark of cancer and a key feature of tumor metabolic reprogramming describes the tendency of tumor cells to rely on glycolysis for energy production, even when sufficient oxygen is available [1, 2]. Lactate was once considered only a byproduct of glycolysis. However, it is now recognized as a versatile regulatory molecule within tumors. Beyond serving as an energy source for tumor growth, lactate plays a role in signal transduction, modulates the pH of the tumor microenvironment, and influences the metabolic behavior of immune cells [3, 4]. Recent advancements in proteomics have revealed lactylation, a novel regulatory mechanism in cancer that utilizes lactate as a substrate for acylation modifications. This discovery provides a new lens for exploring tumor biology, particularly in the context of lactate metabolism [5].
Gene expression is heavily influenced by chromatin remodeling, with dynamic, cell-specific regulatory modifications like histone acetylation and methylation playing a vital role. These reversible changes enable cells to adapt and regulate gene functions in response to environmental stress [6, 7]. Since its discovery, histone lysine lactylation (Kla) has emerged as a distinctive epigenetic modification due to its widespread occurrence, reliance on readily available glycolytic products, and dependence on substrate concentration [8,9,10,11]. Notably, lactylation is not confined to histones, highlighting its broad potential in tumors characterized by the Warburg effect [12, 13].
The interconnected roles of metabolism, epigenetics, and protein modification regulation have opened numerous pathways for cancer treatment [14,15,16]. In the early exploration of lactylation, understanding its relationship with metabolic reprogramming and tumor evolution is crucial. This knowledge lays the groundwork for advancing biological research and translating findings into clinical applications.
This review highlights the role of lactylation modifications in tumor development, progression, and the remodeling of the tumor immune microenvironment. It also discusses existing targets, drugs, and future research directions. By systematically categorizing the molecules involved in lactylation regulation and their associated therapeutic agents, this review aims to offer new insights into leveraging lactylation for early cancer diagnosis and targeted treatments.
Results
Lactate, lactylation, and tumorigenesis
Tumor development is intricately linked to the inactivation of tumor suppressor genes, with dysregulated cellular metabolism standing as a hallmark of cancer [1, 17]. The interaction between unchecked growth signals and tumorigenesis has been a focal point of research. In 2012, Contractor Tet et al. identified the pivotal role of P53 in regulating the Warburg effect. Their study proposed a novel mechanism in which P53 modulates PDK2 activity to shift tumor metabolic preferences, thereby promoting tumorigenesis [18]. Subsequently, classical oncogenes such as PTEN and KRAS were demonstrated to regulate lactate metabolism in tumor cells [19,20,21]. Lactate exhibits diverse pro-carcinogenic effects, including promoting early local tumor invasion by aiding extracellular matrix degradation and enhancing tumor angiogenesis. Additionally, elevated lactate concentrations in the tumor microenvironment influence the activity of T cells and macrophages, contributing to immune evasion during the early stages of tumor development [22,23,24]. Lactylation, a post-translational modification using lactate as a substrate, provides a key regulatory mechanism in early tumorigenesis. In tumor cells, AARS1 detects lactate and drives global lysine lactylation, including the lactylation of P53. This modification impairs the function of p53, suppressing its tumor-suppressive capabilities, and offers a novel perspective on the role of p53 dysfunction in cancer progression [17]. Glycolysis-driven lactylation of the nucleolar protein NCL by the enzyme P300 inhibits alternative splicing associated with the translation termination of downstream MADD. This process ultimately promotes tumor growth through the NCL/MADD/pERK axis, offering a new dimension to the activation of the MAPK signaling pathway in cancer progression [25]. The regulatory role of lactylation in tumorigenesis offers valuable insights and theoretical foundations for early cancer diagnosis. While current methods for detecting lactylation, such as metabolomics and antibody-based techniques, lack systematic clinical studies focused on diagnostics, the potential for leveraging this mechanism as a tool for early cancer detection is highly promising.
Lactylation and tumor evolution
The excessive accumulation of lactate in tumors, driven by the Warburg effect or other metabolic shifts, supplies the substrate needed for intratumoral protein lactylation. This sets the stage for diverse molecule-mediated lactylation regulatory processes. Understanding this mechanism reveals a novel pathway for tumor evolution, extending beyond traditional DNA-level regulation.
Warburg effect and lactic acid-driven epigenetic control
The Warburg effect plays a crucial role in driving tumor cell growth, acidifying and reshaping the tumor microenvironment, and regulating tumor immunity [26]. This metabolic shift alters the availability of various intermediates that directly influence epigenetic modifications. For instance, acetyl-CoA, a key substrate for histone acetylation, experiences level fluctuations that impact global histone acetylation states [27]. Typically, cytoplasmic acetyl-CoA concentrations decrease, while mitochondrial levels remain stable or may slightly increase [28]. In the context of the Warburg effect, increased glycolysis reduces the amount of pyruvate entering the mitochondria. This diminishes the production of acetyl-CoA via the pyruvate dehydrogenase complex (PDC), potentially affecting histone acetylation and, consequently, gene expression [26, 27]. Additionally, lactate accumulation resulting from the Warburg effect significantly impacts the epigenetic regulation of tumor cells. This occurs through mechanisms such as cellular acidification, which affects the activity of histone deacetylases (HDACs), or alterations in the NAD+/NADH ratio, influencing the activity of the NAD+-dependent deacetylase SIRT1. These changes collectively modulate the global epigenetic landscape within tumor cells [29, 30]. Lactylation directly regulates downstream processes and functions as a rapid gene regulatory mechanism, comparable to acetylation. However, while some tumor cells may sustain acetyl-CoA levels through enhanced glutamine metabolism or fatty acid oxidation, lactylation often takes precedence in gene regulation due to its greater substrate availability [31]. Since Zhang et al. first identified lactylation in 2019, the field has advanced significantly, with H3K18La emerging as the most studied histone lactylation modification [32]. In neuroendocrine prostate cancer (NEPC), this modification facilitates cellular plasticity, playing a pivotal role in tumor evolution. The epithelial-mesenchymal transition (EMT) regulator ZEB1 influences H3K18La to enhance the expression of key glycolytic enzymes such as LDHA. This shift alters tumor metabolic preferences and increases chromatin accessibility near neuro-related genes, thereby promoting tumor cell plasticity and adaptability [33]. Research by Yu et al. revealed that H3K18La contributes to tumorigenesis by upregulating YTHDF2, an m6A reader, which promotes the degradation of PER1 and TP53 mRNA [34]. This discovery highlights a novel connection between the Warburg effect and epigenetic regulation, deepening our understanding of how metabolic and epigenetic pathways interact in cancer development.
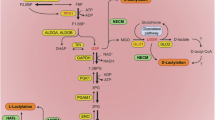
Lactate sensors and lactylation control
We have compiled a summary of potential targets and therapeutic agents aimed at controlling lactylation, presented in Table 1 and Fig. 1. Additionally, Table 2 provides detailed information on those targets and drugs currently in clinical trials, excluding any that have already reached commercialization. This overview offers a comprehensive perspective on the translational progress in this area of research.
Lactylation-related metabolic transport enzymes
Members of the monocarboxylate transporter (MCT) protein family are known not only for their traditional role in transporting monocarboxylates such as lactate across membranes but also for their involvement in lactate sensing and lactylation regulation. MCT1, primarily responsible for lactate uptake, is abundant in oxidative tumor cells, which utilize lactate from the tumor microenvironment as an energy source [35]. MCT1-mediated lactylation plays a key role in stabilizing HIF1A, enhancing KIAA1199 transcription to promote tumor angiogenesis [36]. Additionally, it influences tumor-infiltrating macrophages by driving their polarization toward the M2 phenotype through increased TNFSF9 expression via H3K18La [37]. MCT4 has also been implicated in the regulation of macrophage polarization from M1 to M2 phenotypes via H3K8la in arteriosclerosis, though similar mechanisms in tumors remain unexplored [38]. The MCT1 inhibitor AZD3965 has completed phase I clinical trials for advanced solid tumors and lymphomas, demonstrating tolerability at doses sufficient to achieve target engagement, and offering promise as a therapeutic agent [39].
In addition to MCT family members, sodium-coupled monocarboxylate transporters (SMCTs), such as SMCT1 and SMCT2, also play a role in cellular metabolism by co-transporting lactate along with sodium ions, utilizing the sodium ion concentration gradient [40, 41]. This mechanism parallels that of the sodium-glucose co-transporter (SGLT2), which is crucial in renal glucose reabsorption [42]. The potential connection between SMCTs and lactylation regulation opens an intriguing avenue for future research, providing further insight into the interplay between cellular transport systems and epigenetic modifications. This linkage could advance our understanding of tumor metabolism and therapeutic targets.
Beyond monocarboxylate transporters, glucose transporters have also been implicated in regulating lactylation. GLUT3 was identified as a regulator of protein lactylation in gastric cancer, marking the first report of a glucose transporter influencing intracellular lactylation in oncology [43]. Further research by Alessandra De Leo et al. demonstrated in glioblastomas that GLUT1 expression in glycolytic monocyte-derived macrophages (MDMs) facilitated intracellular lactate-driven histone lactylation near the IL-10 gene. This process led to increased IL-10 accumulation in the tumor microenvironment, suppressing T-cell activity [44]. These findings provide a novel perspective on the traditional roles of glycolysis-related transporters, highlighting their contributions to tumor metabolism and immune modulation.
Lactylation-related metabolic enzymes
Key glycolysis-related enzymes have long been pivotal in studying tumor metabolism and exploring translational applications. Protein lactylation, driven by lactate accumulation, introduces a novel mechanism for regulating tumor functions through metabolic enzymes.
PKM2, a pyruvate kinase isozyme, is highly expressed in rapidly proliferating cancer cells, catalyzing the final step of glycolysis to produce pyruvate and ATP. PKM2 alternates between an active tetrameric form and a less active dimeric form [45]. In glioblastomas, the interaction of ALDH1A3 with PKM2 promotes tetramer formation, leading to lactate accumulation in cancer stem cells. This, in turn, triggers lactylation-induced nuclear translocation of XRCC1, enhancing DNA repair and potentially contributing to tumor resistance to radiotherapy and chemotherapy [13]. Lactylation also impacts non-glycolytic metabolism. Tumor cells, often exposed to oxidative stress, rely on G6PD to generate NADPH, which is crucial for managing oxidative stress and supporting proliferation through the pentose phosphate pathway (PPP). In HPV-related cervical cancer, the HPV16 E6E7 protein inhibits lactylation of G6PD at the K45 site, promoting G6PD dimerization and enhancing its activity. This activation of the PPP fuels rapid tumor growth [46]. RRx-001, a G6PD inhibitor, has been under clinical investigation since 2011 for advanced malignancies, including colorectal cancer, lymphoma, and head and neck cancer, with promising outcomes [47]. Its latest phase III clinical trial targets small-cell lung cancer and is currently recruiting, offering potential advancements in cancer therapy.
Lactyltransferases
P300 (EP300) was initially identified as a transcriptional coactivator that enhances gene expression by interacting with various transcription factors. In 2019, Zhang et al. proposed its potential role as a lactyltransferase [32]. Subsequent studies confirmed P300’s lactyltransferase activity in cancers such as pancreatic ductal adenocarcinoma (PDAC) and intrahepatic cholangiocarcinoma, implicating it in lactylation regulation [31, 48]. However, P300 is not exclusive to lactylation—it also catalyzes other acyl modifications. P300 serves as a critical integrator of metabolic and transcriptional signals. By regulating acetylation with interactions involving MYC, AKT, and TGF-β, P300 drives epigenetic changes that promote tumor growth and progression [49,50,51]. The proportion or preference of P300-mediated protein lactylation and acetylation, as well as their impact on regulatory networks, await further research and reporting. Understanding this crosstalk could open new avenues for targeted cancer therapies. AARS1 was the first identified lactyltransferase and plays a critical role in sensing intracellular lactate. It translocates to the nucleus, where it lactylates and activates the YAP-TEAD complex, a key component of the Hippo pathway. This process is subject to positive feedback regulation by YAP-TEAD itself [12]. Similarly, the acetyltransferase KAT5 (TIP60), known for facilitating histone acetylation, also mediates lactylation. KAT5/TIP60-driven lactylation of PIK3C3/VPS34 at lysine residues 356 and 781 enhances its interaction with BECN1, ATG14, and UVRAG, promoting autophagy that may contribute to cancer progression [52]. Research on lactylation-specific “writers” remains in its infancy, and further studies are needed to elucidate their precise roles and mechanisms in cancer biology.
De-lactylases
The HDAC (histone deacetylase) family was originally identified for its role in regulating chromatin structure and gene expression by removing acetyl groups from histones [53]. Building on their discovery of P300 as a lactyl “writer,” Professor Zhao Yingming’s team reported that common deacetylases, including HDAC1-3 and SIRT1-3, can remove lactylation modifications, identifying them as potential lactyl “erasers.” They further proposed the specificity of lactylation sites and the possibility of other lactyl group erasers yet to be discovered [54]. Class I HDACs (HDAC1-3) were confirmed by Zhao’s team as the most effective lysine de-lactylases in vitro. Despite this, reports linking HDACs to the regulation of lactylation in cancer are scarce. Notably, HDAC inhibitors, such as tucidinostat, are already approved for cancer treatment, suggesting potential for therapeutic exploration in targeting lactylation pathways via HDAC modulation. Further research is needed to better understand the role of HDACs in lactylation regulation and their implications in cancer therapy [55].
Vorinostat (SAHA, Zolinza) was the first HDAC inhibitor approved by the FDA in 2006, achieving a total remission rate of 30% in patients with recurrent or refractory cutaneous T-cell lymphoma (CTCL) [56]. Since then, HDAC inhibitors have undergone significant advancements, demonstrating efficacy in select conditions. However, their broader application, particularly as monotherapies for solid tumors, has remained limited [57]. Research into HDAC regulation of lactylation may pave the way for a new generation of HDAC inhibitors with enhanced specificity and efficacy.
SIRTs (NAD+-dependent deacetylases) also play a role in tumor suppression. SIRT2 and SIRT3 have been reported to inhibit tumor progression by de-lactylating non-histone proteins [58, 59]. Despite the absence of lactyl group-specific de-lactylases, the growing body of research on deacetylases suggests significant potential for identifying “erasers” of lactylation modifications.
Further investigation into these acyl modification “erasers” could deepen our understanding of their regulatory mechanisms in cancer and inspire new strategies for clinical applications. By bridging insights from lactylation biology and therapeutic development, these studies hold promise for more targeted and effective cancer treatments.
Lactylation and tumor microenvironment remodeling
Lactylation utilizes the metabolic products of glucose, namely lactate, and ATP, to covalently modify proteins, achieving regulatory effects. This mechanism shares similarities with ATP-dependent phosphorylation, suggesting that lactylation may serve as a convenient and widespread regulatory method. Over time, numerous key proteins and prominent cell types have been identified as participants in this novel process.
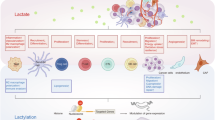
Current research predominantly focuses on the role of lactylation in mediating epigenetic changes in immune cells under lactate stress within the tumor microenvironment. However, this raises intriguing questions about whether other critical intracellular targets are influenced by lactate and operate through lactylation. Exploring these mechanisms further could uncover additional pathways and targets affected by this regulatory mode, expanding our understanding of lactylation’s role in cellular and tumor biology (Fig. 2).
Lactylation in immune cells: epigenetic control
The Warburg effect in tumors drives lactate accumulation in the tumor microenvironment, reshaping immune cell metabolism and perpetuating a cycle of malignant tumor behavior, immune cell remodeling, and environmental immunosuppression [3]. Tumor-intrinsic PI3K pathways amplify lactate production and accumulation, exacerbating this cycle. Under metabolic stress, tumor-associated macrophages (TAMs) exhibit increased histone lactylation at H3K18, reducing their anti-cancer phagocytic activity [21]. Jia Xiong et al. found that lactate accumulation induces upregulation of METTL3 in tumor-infiltrating myeloid cells (TIMs) via H3K18la. This upregulation promotes m6A modification of Jak1 mRNA, enhancing protein translation and activating the JAK-STAT pathway, which further supports the immunosuppressive role of TIMs [60]. Similarly, within the STAT family, STAT5 has been shown to elevate PD-L1 expression on leukemia cells through histone lactylation, suppressing normal T-cell activation and contributing to immune evasion. These findings underscore the critical role of lactylation in epigenetically modulating immune responses within the tumor microenvironment [61].
Immunosuppressive signaling enhancement
Lactylation at key signaling sites within the tumor microenvironment amplifies immunosuppressive signaling, undermining normal tumor immunity. For instance, lactate-driven lactylation of MOESIN enhances the production of regulatory T cells (Tregs) in the tumor microenvironment. This modification improves MOESIN’s interaction with the TGF-β receptor, activating the classical SMAD3 signaling pathway and fostering immune suppression [62]. In innate immunity, the cGAS protein is essential for immune surveillance, detecting mitochondrial DNA (mtDNA) or chromosomal DNA fragments. However, lactylation of cGAS by AARS2 inactivates cGAS, suppressing the synthesis of cGAMP and impairing the innate immune response. Studies show that blocking monocarboxylate transporter 1 (MCT1) to inhibit L-lactate transport can prevent cGAS lactylation, thereby restoring innate immunity [63]. While cGAS lactylation has not yet been reported in oncology, these findings expand our understanding of the interplay between classic immune pathways and lactylation. This knowledge offers valuable insights and a theoretical framework for developing innovative immunotherapeutic strategies targeting lactylation-driven immune dysregulation.
Concluding remarks
Lactylation, a novel post-translational modification (PTM) utilizing lactyl residues, has significantly advanced our understanding of lactate’s role in tumor initiation and progression. This modification not only impacts histones by altering chromatin structure and DNA accessibility to regulate gene expression but also broadly influences the activity of key molecules, akin to phosphorylation. The close relationship between lactate concentration and lactylation establishes a direct link between metabolic dysregulation in tumors and other cancer hallmarks, such as sustained proliferation signals and immune evasion [1, 64].
Research on lactylation in tumors builds upon foundational discoveries in tumor metabolism, paving the way for clinical translation based on metabolic and lactylation regulatory mechanisms. While concerns persist about the specificity of lactylation-related “writers” and “erasers” posing challenges for targeted therapies, the identification of AARS1/2 as specific lactylation writers offer hope for precise regulation and therapeutic innovation.
The translational challenges are considerable, as many inhibitors and drugs focus on enzymes like LDH, MCTs, and PKM. These enzymes, similar to lactylation “writers” and “erasers,” also regulate other critical metabolic reactions, necessitating careful evaluation to balance potential benefits and risks in tumor patients. For instance, while alanine has been identified as a competitive inhibitor of AARS1, gaps in our understanding of its broader implications remain significant [65].
Although lactate accumulation is a necessary condition and environmental feature for lactylation, the potential influence and regulation of other specific tumor microenvironments, such as hypoxia and inflammatory conditions, cannot be excluded. This part of the puzzle urgently awaits further research for clarification. Moreover, different cancer types exhibit varying metabolic preferences. Tumors originating from tissues highly reliant on glycolytic metabolism or those from tissues that minimally utilize glycolysis for energy may possess distinct lactylation characteristics. Most studies suggest that histone lactylation supports tumor progression, often promoting tumor growth. However, whether lactylation is inherently pro-carcinogenic or context-dependent requires further exploration. Current research has only touched the surface of lactylation’s potential as a target in cancer therapy. It is not merely an epigenetic pathway and should not be approached simplistically as such.
Further investigation into lactylation’s role in tumor initiation and the regulation of the tumor immune microenvironment holds promise for breakthroughs in cancer diagnostics and treatment. In particular, lactylation detection could enable early diagnosis, and immunomodulatory drugs that inhibit lactylation may emerge as effective therapeutic strategies. These advancements could redefine the landscape of cancer care by integrating metabolic and immunological perspectives.
References
Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46.
Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37.
Apostolova P, Pearce EL. Lactic acid and lactate: revisiting the physiological roles in the tumor microenvironment. Trends Immunol. 2022;43:969–77.
Magistretti PJ, Allaman I. Lactate in the brain: from metabolic end-product to signalling molecule. Nat Rev Neurosci. 2018;19:235–49.
Yang Z, Yan C, Ma J, Peng P, Ren X, Cai S, et al. Lactylome analysis suggests lactylation-dependent mechanisms of metabolic adaptation in hepatocellular carcinoma. Nat Metab. 2023;5:61–79.
Stefanska B, Karlic H, Varga F, Fabianowska-Majewska K, Haslberger A. Epigenetic mechanisms in anti-cancer actions of bioactive food components-the implications in cancer prevention. Br J Pharmacol. 2012;167:279–97.
Yen CY, Huang HW, Shu CW, Hou MF, Yuan SS, Wang HR, et al. DNA methylation, histone acetylation and methylation of epigenetic modifications as a therapeutic approach for cancers. Cancer Lett. 2016;373:185–92.
Li F, Zhang H, Huang Y, Li D, Zheng Z, Xie K, et al. Single-cell transcriptome analysis reveals the association between histone lactylation and cisplatin resistance in bladder cancer. Drug Resist Updat. 2024;73:101059.
Li W, Zhou C, Yu L, Hou Z, Liu H, Kong L, et al. Tumor-derived lactate promotes resistance to bevacizumab treatment by facilitating autophagy enhancer protein RUBCNL expression through histone H3 lysine 18 lactylation (H3K18la) in colorectal cancer. Autophagy. 2024;20:114–30.
Niu Z, Chen C, Wang S, Lu C, Wu Z, Wang A, et al. HBO1 catalyzes lysine lactylation and mediates histone H3K9la to regulate gene transcription. Nat Commun. 2024;15:3561.
Zhang Y, Song H, Li M, Lu P. Histone lactylation bridges metabolic reprogramming and epigenetic rewiring in driving carcinogenesis: Oncometabolite fuels oncogenic transcription. Clin Transl Med. 2024;14:e1614.
Ju J, Zhang H, Lin M, Yan Z, An L, Cao Z, et al. The alanyl-tRNA synthetase AARS1 moonlights as a lactyltransferase to promote YAP signaling in gastric cancer. J Clin Investig. 2024;134:e174587.
Li G, Wang D, Zhai Y, Pan C, Zhang J, Wang C, et al. Glycometabolic reprogramming-induced XRCC1 lactylation confers therapeutic resistance in ALDH1A3-overexpressing glioblastoma. Cell Metab. 2024;36:1696–1710.e10.
Stine ZE, Schug ZT, Salvino JM, Dang CV. Targeting cancer metabolism in the era of precision oncology. Nat Rev Drug Discov. 2022;21:141–62.
Sun T, Liu Z, Yang Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol Cancer. 2020;19:146.
Cao J, Yan Q. Cancer epigenetics, tumor immunity, and immunotherapy. Trends Cancer. 2020;6:580–592.
Marshall CJ. Tumor suppressor genes. Cell. 1991;64:313–26.
Contractor T, Harris CR. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 2012;72:560–7.
Hutton JE, Wang X, Zimmerman LJ, Slebos RJ, Trenary IA, Young JD, et al. Oncogenic KRAS and BRAF drive metabolic reprogramming in colorectal cancer. Mol Cell Proteomics. 2016;15:2924–38.
Byun JK, Park M, Yun JW, Lee J, Kim JS, Cho SJ, et al. Oncogenic KRAS signaling activates mTORC1 through COUP-TFII-mediated lactate production. EMBO Rep. 2019;20:e47451.
Chaudagar K, Hieromnimon HM, Khurana R, Labadie B, Hirz T, Mei S, et al. Reversal of lactate and PD-1-mediated Macrophage Immunosuppression Controls Growth of PTEN/p53-deficient prostate cancer. Clin Cancer Res. 2023;29:1952–68.
Végran F, Boidot R, Michiels C, Sonveaux P, Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-κB/IL-8 pathway that drives tumor angiogenesis. Cancer Res. 2011;71:2550–60.
Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473.
Qian Y, Galan-Cobo A, Guijarro I, Dang M, Molkentine D, Poteete A, et al. MCT4-dependent lactate secretion suppresses antitumor immunity in LKB1-deficient lung adenocarcinoma. Cancer Cell. 2023;41:1363–1380.e7.
Yang L, Niu K, Wang J, Shen W, Jiang R, Liu L, et al. Nucleolin lactylation contributes to intrahepatic cholangiocarcinoma pathogenesis via RNA splicing regulation of MADD. J Hepatol. 2024;81:651–666.
Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–8.
Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80.
Etchegaray JP, Mostoslavsky R. Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol Cell. 2016;62:695–711.
Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–60.
Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ, et al. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012;40:4794–803.
Yang C, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell. 2014;56:414–24.
Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–80.
Wang D, Du G, Chen X, Wang J, Liu K, Zhao H, et al. Zeb1-controlled metabolic plasticity enables remodeling of chromatin accessibility in the development of neuroendocrine prostate cancer. Cell Death Differ. 2024;31:779–91.
Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X, et al. Histone lactylation drives oncogenesis by facilitating m(6)A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021;22:85.
Felmlee MA, Jones RS, Rodriguez-Cruz V, Follman KE, Morris ME. Monocarboxylate transporters (SLC16): function, regulation, and role in health and disease. Pharmacol Rev. 2020;72:466–85.
Luo Y, Yang Z, Yu Y, Zhang P. HIF1α lactylation enhances KIAA1199 transcription to promote angiogenesis and vasculogenic mimicry in prostate cancer. Int J Biol Macromol. 2022;222:2225–2243.
Li M, Sun P, Tu B, Deng G, Li D, He W. Hypoxia conduces the glioma progression by inducing M2 macrophage polarization via elevating TNFSF9 level in a histone-lactylation-dependent manner. Am J Physiol Cell Physiol. 2024;327:C487–c504.
Zhang Y, Jiang H, Dong M, Min J, He X, Tan Y, et al. Macrophage MCT4 inhibition activates reparative genes and protects from atherosclerosis by histone H3 lysine 18 lactylation. Cell Rep. 2024;43:114180.
Halford S, Veal GJ, Wedge SR, Payne GS, Bacon CM, Sloan P, et al. A phase I dose-escalation study of AZD3965, an oral monocarboxylate transporter 1 inhibitor, in patients with advanced cancer. Clin Cancer Res. 2023;29:1429–39.
Han L, Qu Q, Aydin D, Panova O, Robertson MJ, Xu Y, et al. Structure and mechanism of the SGLT family of glucose transporters. Nature. 2022;601:274–9.
Pucino V, Certo M, Bulusu V, Cucchi D, Goldmann K, Pontarini E, et al. Lactate buildup at the site of chronic inflammation promotes disease by inducing CD4(+) T cell metabolic rewiring. Cell Metab. 2019;30:1055–1074.e8.
Ferrannini E. Sodium-glucose co-transporters and their inhibition: clinical physiology. Cell Metab. 2017;26:27–38.
Yang H, Yang S, He J, Li W, Zhang A, Li N, et al. Glucose transporter 3 (GLUT3) promotes lactylation modifications by regulating lactate dehydrogenase A (LDHA) in gastric cancer. Cancer Cell Int. 2023;23:303.
De Leo A, Ugolini A, Yu X, Scirocchi F, Scocozza D, Peixoto B, et al. Glucose-driven histone lactylation promotes the immunosuppressive activity of monocyte-derived macrophages in glioblastoma. Immunity. 2024;57:1105–1123.e8.
Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–47.
Meng Q, Zhang Y, Sun H, Yang X, Hao S, Liu B, et al. Human papillomavirus-16 E6 activates the pentose phosphate pathway to promote cervical cancer cell proliferation by inhibiting G6PD lactylation. Redox Biol. 2024;71:103108.
Chen Y, He H, Lin B, Chen Y, Deng X, Jiang W, et al. RRx-001 ameliorates inflammatory diseases by acting as a potent covalent NLRP3 inhibitor. Cell Mol Immunol. 2021;18:1425–36.
Li F, Si W, Xia L, Yin D, Wei T, Tao M, et al. Positive feedback regulation between glycolysis and histone lactylation drives oncogenesis in pancreatic ductal adenocarcinoma. Mol Cancer. 2024;23:90.
Brand M, Clayton J, Moroglu M, Schiedel M, Picaud S, Bluck JP, et al. Controlling intramolecular interactions in the design of selective, high-affinity ligands for the CREBBP bromodomain. J Med Chem. 2021;64:10102–23.
Wang J, Wu M, Zheng D, Zhang H, Lv Y, Zhang L, et al. Garcinol inhibits esophageal cancer metastasis by suppressing the p300 and TGF-β1 signaling pathways. Acta Pharmacol Sin. 2020;41:82–92.
Dou C, Liu Z, Tu K, Zhang H, Chen C, Yaqoob U, et al. P300 Acetyltransferase mediates stiffness-induced activation of hepatic stellate cells into tumor-promoting myofibroblasts. Gastroenterology. 2018;154:2209–2221.e14.
Sun W, Jia M, Feng Y, Cheng X. Lactate is a bridge linking glycolysis and autophagy through lactylation. Autophagy. 2023;19:3240–1.
Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–11.
Moreno-Yruela C, Zhang D, Wei W, Bæk M, Liu W, Gao J, et al. Class I histone deacetylases (HDAC1-3) are histone lysine delactylases. Sci Adv. 2022;8:eabi6696.
Fan W, Zeng S, Wang X, Wang G, Liao D, Li R, et al. A feedback loop driven by H3K9 lactylation and HDAC2 in endothelial cells regulates VEGF-induced angiogenesis. Genome Biol. 2024;25:165.
Mann BS, Johnson JR, He K, Sridhara R, Abraham S, Booth BP, et al. Vorinostat for treatment of cutaneous manifestations of advanced primary cutaneous T-cell lymphoma. Clin Cancer Res. 2007;13:2318–22.
Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci. 2017;18:1414.
Jin J, Bai L, Wang D, Ding W, Cao Z, Yan P, et al. SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth. EMBO Rep. 2023;24:e56052.
Zu H, Li C, Dai C, Pan Y, Ding C, Sun H, et al. SIRT2 functions as a histone delactylase and inhibits the proliferation and migration of neuroblastoma cells. Cell Discov. 2022;8:54.
Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82:1660–1677.e10.
Huang ZW, Zhang XN, Zhang L, Liu LL, Zhang JW, Sun YX, et al. STAT5 promotes PD-L1 expression by facilitating histone lactylation to drive immunosuppression in acute myeloid leukemia. Signal Transduct Target Ther. 2023;8:391.
Gu J, Zhou J, Chen Q, Xu X, Gao J, Li X, et al. Tumor metabolite lactate promotes tumorigenesis by modulating MOESIN lactylation and enhancing TGF-β signaling in regulatory T cells. Cell Rep. 2022;39:110986.
Li H, Liu C, Li R, Zhou L, Ran Y, Yang Q, et al. AARS1 and AARS2 sense L-lactate to regulate cGAS as global lysine lactyltransferases. Nature. 2024;634:1229–37.
Wang J, Wang Z, Wang Q, Li X, Guo Y. Ubiquitous protein lactylation in health and diseases. Cell Mol Biol Lett. 2024;29:23.
Zong Z, Xie F, Wang S, Wu X, Zhang Z, Yang B, et al. Alanyl-tRNA synthetase, AARS1, is a lactate sensor and lactyltransferase that lactylates p53 and contributes to tumorigenesis. Cell. 2024;187:2375–2392.e33.
Bader DA, Hartig SM, Putluri V, Foley C, Hamilton MP, Smith EA, et al. Mitochondrial pyruvate import is a metabolic vulnerability in androgen receptor-driven prostate cancer. Nat Metab. 2019;1:70–85.
Dong L, Lu D, Chen R, Lin Y, Zhu H, Zhang Z, et al. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell. 2022;40:70–87.e15.
Ma XM, Geng K, Wang P, Jiang Z, Law BY, Xu Y. MCT4-dependent lactate transport: a novel mechanism for cardiac energy metabolism injury and inflammation in type 2 diabetes mellitus. Cardiovasc Diabetol. 2024;23:96.
Li Y, Tang S, Shi X, Lv J, Wu X, Zhang Y, et al. Metabolic classification suggests the GLUT1/ALDOB/G6PD axis as a therapeutic target in chemotherapy-resistant pancreatic cancer. Cell Rep Med. 2023;4:101162.
Kanakkanthara A, Kurmi K, Ekstrom TL, Hou X, Purfeerst ER, Heinzen EP, et al. BRCA1 deficiency upregulates NNMT, which reprograms metabolism and sensitizes ovarian cancer cells to mitochondrial metabolic targeting agents. Cancer Res. 2019;79:5920–9.
Olszewski K, Barsotti A, Feng XJ, Momcilovic M, Liu KG, Kim JI, et al. Inhibition of glucose transport synergizes with chemical or genetic disruption of mitochondrial metabolism and suppresses TCA cycle-deficient tumors. Cell Chem Biol. 2022;29:423–435.e10.
Leng L, Yuan Z, Pan R, Su X, Wang H, Xue J, et al. Microglial hexokinase 2 deficiency increases ATP generation through lipid metabolism leading to β-amyloid clearance. Nat Metab. 2022;4:1287–305.
Shigeta K, Hasegawa M, Hishiki T, Naito Y, Baba Y, Mikami S, et al. IDH2 stabilizes HIF-1α-induced metabolic reprogramming and promotes chemoresistance in urothelial cancer. Embo j. 2023;42:e110620.
Zawistowski JS, Bevill SM, Goulet DR, Stuhlmiller TJ, Beltran AS, Olivares-Quintero JF, et al. Enhancer remodeling during adaptive bypass to MEK inhibition is attenuated by pharmacologic targeting of the P-TEFb complex. Cancer Discov. 2017;7:302–21.
Edwards DS, Maganti R, Tanksley JP, Luo J, Park JJH, Balkanska-Sinclair E, et al. BRD4 prevents R-loop formation and transcription-replication conflicts by ensuring efficient transcription elongation. Cell Rep. 2020;32:108166.
Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–65.
Xie J, Wang Z, Fan W, Liu Y, Liu F, Wan X, et al. Targeting cancer cell plasticity by HDAC inhibition to reverse EBV-induced dedifferentiation in nasopharyngeal carcinoma. Signal Transduct Target Ther. 2021;6:333.
Pei Y, Liu KW, Wang J, Garancher A, Tao R, Esparza LA, et al. HDAC and PI3K antagonists cooperate to inhibit growth of MYC-driven medulloblastoma. Cancer Cell. 2016;29:311–23.
Butler LM, Webb Y, Agus DB, Higgins B, Tolentino TR, Kutko MC, et al. Inhibition of transformed cell growth and induction of cellular differentiation by pyroxamide, an inhibitor of histone deacetylase. Clin Cancer Res. 2001;7:962–70.
Gao Y, Nihira NT, Bu X, Chu C, Zhang J, Kolodziejczyk A, et al. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat Cell Biol. 2020;22:1064–75.
Adeegbe DO, Liu Y, Lizotte PH, Kamihara Y, Aref AR, Almonte C, et al. Synergistic immunostimulatory effects and therapeutic benefit of combined histone deacetylase and bromodomain inhibition in non-small cell lung cancer. Cancer Discov. 2017;7:852–67.
Wang Z, Hausmann S, Lyu R, Li TM, Lofgren SM, Flores NM, et al. SETD5-coordinated chromatin reprogramming regulates adaptive resistance to targeted pancreatic cancer therapy. Cancer Cell. 2020;37:834–849.e13.
Feng YQ, Liu X, Zuo N, Yu MB, Bian WM, Han BQ, et al. NAD(+) precursors promote the restoration of spermatogenesis in busulfan-treated mice through inhibiting Sirt2-regulated ferroptosis. Theranostics. 2024;14:2622–36.
Hamaidi I, Zhang L, Kim N, Wang MH, Iclozan C, Fang B, et al. Sirt2 inhibition enhances metabolic fitness and effector functions of tumor-reactive T cells. Cell Metab. 2020;32:420–436.e12.
Qian K, Tang J, Ling YJ, Zhou M, Yan XX, Xie Y, et al. Exogenous NADPH exerts a positive inotropic effect and enhances energy metabolism via SIRT3 in pathological cardiac hypertrophy and heart failure. EBioMedicine. 2023;98:104863.
Acknowledgements
We would like to express our gratitude to Biorender for providing us with beautiful templates for our scientific illustrations and supporting us in the publication process.
Author information
Authors and Affiliations
Contributions
YS and ZYS were responsible for collecting and arranging references, writing, and revising the manuscript. JFF, ZLW, and GRZ provided ideas and helped organize the structure and main content of this manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sui, Y., Shen, Z., Wang, Z. et al. Lactylation in cancer: metabolic mechanism and therapeutic strategies. Cell Death Discov. 11, 68 (2025). https://doi.org/10.1038/s41420-025-02349-4
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41420-025-02349-4
This article is cited by
-
Emerging role of histone and non-histone lactylation in metabolic reprogramming of female-specific malignancies
World Journal of Surgical Oncology (2025)
-
Lactate and lactylation: molecular insights into histone and non-histone lactylation in tumor progression, tumor immune microenvironment, and therapeutic strategies
Biomarker Research (2025)