Abstract
Tobacco use, including both traditional and electronic cigarettes, profoundly alters host–microbiota interactions, contributing to the pathogenesis of various systemic diseases. Smoking-induced microbial dysbiosis impacts multiple anatomical sites, including the oral cavity, respiratory tract, and gastrointestinal system, driving disease progression through mechanisms such as immune modulation, chronic inflammation, and metabolic dysregulation. This review examines the disruption of microbial ecosystems by smoking, with a focus on the imbalance between beneficial and pathogenic microorganisms. In the oral cavity, smoking is strongly linked to diseases such as periodontitis and oral cancer, marked by shifts in microbial diversity and functional profiles. Similar dysbiotic changes are observed in the respiratory and gastrointestinal systems, where smoking impairs mucosal immunity, increases oxidative stress, and compromises barrier integrity, thereby enhancing susceptibility to chronic diseases. Additionally, the review addresses the challenges in establishing causality between microbial changes and disease outcomes, emphasizing the need for more comprehensive research utilizing multi-omics approaches and longitudinal studies. By exploring the potential for microbiota-based interventions, this review underscores the critical role of microbial dysbiosis in smoking-related health risks, providing valuable insights for the development of targeted therapeutic strategies to mitigate the global health burden of tobacco use.
Similar content being viewed by others
Facts
-
Causal link between microbial dysbiosis and disease outcomes: The relationship between smoking-induced microbial imbalances (e.g., Fusobacterium, Prevotella enrichment) and disease progression (e.g., periodontitis, COPD, colorectal cancer) remains correlative. Mechanistic studies are needed to determine whether dysbiosis directly drives pathology or is a secondary effect of inflammation or epithelial damage.
-
Electronic vs. traditional cigarettes: Comparative impacts of electronic cigarettes (ECs) and combustible cigarettes on microbial communities—such as EC-specific enrichment of Veillonella and suppression of Porphyromonas—warrant further investigation to clarify distinct disease risks (e.g., oral preneoplasia, periodontal inflammation).
-
Reversibility post-cessation: Evidence suggests partial restoration of oral and gut microbiota after smoking cessation (e.g., Neisseria recovery in saliva, Lactobacillaceae increase with modified-risk products), but long-term ecological resilience and its health implications require longitudinal human cohort studies.
-
Gut–lung axis modulation: Smoking-induced disruptions in short-chain fatty acid (SCFA) production and bile acid metabolism (e.g., taurodeoxycholic acid elevation) may link gut dysbiosis to pulmonary inflammation and cancer, emphasizing the need for multi-omics integration to decode systemic microbial cross-talk.
Introduction
Globally, over 1 billion individuals smoke cigarettes, contributing significantly to the global health burden [1]. Chronic smoking is strongly associated with increased morbidity and a marked reduction in life expectancy, with an estimated loss of up to a decade of life in developing countries [2, 3]. Cigarette smoking (CS) is a well-established risk factor for a wide range of systemic diseases, including cardiovascular disorders, respiratory diseases, diabetes, inflammatory bowel diseases, and various cancers [4, 5]. Smoking exerts direct toxicological effects on the cardiovascular system, contributing to conditions such as atherosclerosis and coronary artery disease, and is a leading cause of chronic obstructive pulmonary disease (COPD) [6, 7]. Additionally, smoking is strongly linked to several cancers, including those of the oral cavity, upper digestive tract, lung, bladder, kidney, and liver. These associations are primarily driven by mechanisms such as the induction of somatic mutations, oxidative stress, and epigenetic modifications, which disrupt cellular function and promote disease progression. Importantly, smoking cessation can substantially reduce these health burdens, leading to significant improvements in overall health outcomes [8, 9].
Beyond its direct systemic toxicological effects, smoking exerts a profound impact on host-associated microbial communities across multiple body sites (Fig. 1). The human microbiota, composed of bacteria, fungi, archaea, and other microorganisms, plays a critical role in regulating immune responses, metabolism, and defense against harmful pathogens. Disruptions to this microbial ecosystem—termed dysbiosis—are implicated in a wide array of diseases [10,11,12]. Smoking has been shown to induce shifts in the microbiota across several body niches, including the oral cavity, respiratory tract, and gastrointestinal system. These microbial alterations contribute to the onset and progression of systemic diseases through various mechanisms, including immune modulation, chronic inflammation, metabolic dysregulation, oxidative stress, epigenetic modifications, and impaired barrier function.
Cigarette smoking disrupts the homeostasis of host-associated microbiota, leading to microbial dysbiosis that affects multiple organ systems. The oral, respiratory, and gastrointestinal tracts—each directly or indirectly exposed to tobacco smoke—exhibit distinct yet interconnected microbial alterations. In the oral cavity, dysbiosis contributes to periodontitis, dental caries, recurrent aphthous stomatitis (RAS), oral malodor, and oral squamous cell carcinoma (OSCC). In the respiratory tract, microbial imbalance exacerbates chronic obstructive pulmonary disease (COPD), asthma, acute respiratory distress syndrome (ARDS), and lung adenocarcinoma (LUAD). In the gastrointestinal tract, smoking-induced dysbiosis compromises mucosal barrier integrity, disrupts immune and metabolic homeostasis, and promotes cardiovascular disease, hepatic dysfunction, inflammatory bowel disease (IBD), and colorectal cancer (CRC). The red arrows indicate the bidirectional crosstalk between microbial dysbiosis and systemic disorders, highlighting how tobacco-induced microbial perturbations propagate through mucosal and immune pathways to influence distal organs. Created in https://BioRender.com.
In the oral cavity, smoking-induced dysbiosis plays a significant role in the development of oral diseases, such as periodontitis, by disrupting the balance of oral microbiota and promoting pathogenic species that drive inflammation and tissue destruction [13, 14]. In the respiratory tract, smoking exacerbates inflammation, leading to increased mucus production, impaired ciliary function, and airway remodeling, which worsen the pathophysiology of respiratory diseases [15, 16]. Within the gastrointestinal system, smoking alters several physiological functions, including the production of mucins, which are critical for mucosal protection, as well as the integrity of tight junctions in the small intestine, which compromises gut barrier function. This disruption can facilitate the translocation of harmful pathogens and toxins, further exacerbating inflammation [17, 18]. Moreover, smoking increases oxidative stress by inducing the production of reactive oxygen species (ROS), which directly damage cellular structures such as lipids, proteins, and DNA, contributing to tissue inflammation, cellular dysfunction, and the progression of chronic diseases. These oxidative effects are compounded by the immune dysregulation caused by microbial alterations [19, 20]. Collectively, these microbial, physiological, and oxidative changes triggered by smoking may amplify its detrimental impact on health, exacerbating a wide range of diseases and impairing the body’s ability to repair and protect itself.
Recent advances in metagenomic sequencing and other omics technologies have provided valuable insights into the specific microbial alterations associated with smoking-related diseases. These technologies have enabled the identification of distinct microbial signatures linked to various smoking-induced pathologies [21,22,23]. However, a fundamental challenge remains in determining whether these microbial changes are causally involved in disease development or if they represent secondary consequences of the pathological processes. Although strong correlations between smoking, pathophysiological outcomes, and microbial dysbiosis have been observed in both human and animal models, these associations alone do not establish causality. Further studies are essential to elucidate the mechanisms underlying these microbial shifts and to determine their role in disease onset and progression. Clarifying these relationships is critical for identifying potential therapeutic targets and developing strategies to mitigate the health impacts of smoking.
This review aims to provide a comprehensive overview of the impact of smoking on the microbiota across the oral, respiratory, and gastrointestinal tracts, and examines the crucial role these microbial alterations play in the pathogenesis of smoking-related diseases. By exploring the systemic effects of smoking on microbial communities, we discuss how these shifts contribute to the onset and exacerbation of a broad range of diseases, including cancer, cardiovascular conditions, and metabolic disorders. Furthermore, we highlight the mechanisms linking smoking-induced microbial dysbiosis to disease, emphasizing how disruptions in the microbiota of key organ systems can significantly influence disease pathophysiology. We also address the limitations of current research, particularly the challenge of distinguishing causality from correlation, and propose future research directions aimed at elucidating the mechanistic pathways involved. By investigating the microbiome’s role in smoking-related health risks, this review offers a novel perspective on potential therapeutic interventions, identifying promising avenues for future research that could lead to more effective strategies for mitigating the harmful effects of smoking on human health.
Literature search methodology
This article is designed as a narrative review aiming to integrate current evidence on smoking-induced microbial dysbiosis and its systemic implications. To ensure a comprehensive and evidence-based synthesis, a literature search was conducted across PubMed, Scopus, and Web of Science databases. Publications from January 2010 to May 2025 were retrieved, with a particular emphasis on studies published within the past 5 years to capture the most recent advances. The search employed combinations of the following keywords: “cigarette”, “microbiome”, “dysbiosis”, “cancer”, “systemic diseases”, “COPD”, “periodontal disease”, and “diabetes”. Only peer-reviewed articles published in English were included. Eligible studies investigated the effects of tobacco exposure on microbial composition and its association with local or systemic pathophysiological outcomes, including inflammatory, metabolic, and neoplastic processes. Reviews, commentaries, conference abstracts, and studies not directly addressing tobacco-related microbial alterations were excluded. Additionally, reference lists of relevant publications were manually screened to identify supplementary studies. This comprehensive strategy aimed to provide an up-to-date and balanced synthesis of current evidence while minimizing potential selection bias.
Overview of human microbiota
To understand how smoking disrupts host–microbe interactions, it is first necessary to outline the composition and function of the human microbiota. Cigarette smoke exerts distinct effects across the oral, respiratory, and gastrointestinal systems, each of which harbors unique microbial ecosystems essential for maintaining physiological homeostasis. A clear overview of these communities provides the foundation for interpreting how smoking-induced perturbations translate into local and systemic disease processes.
The human microbiota constitutes a vast and intricate assembly of microorganisms that inhabit various regions of the human body, playing an indispensable role in maintaining overall health and supporting critical physiological functions. This diverse community, which includes bacteria, viruses, fungi, and archaea, has co-evolved with humans over millennia, forming complex ecosystems that are essential for processes such as nutrient absorption, immune modulation, and pathogen defense [24,25,26]. Microorganisms within the microbiota assist in the digestion of food, the synthesis of essential vitamins, and the fermentation of fibers into short-chain fatty acids, which are important energy sources for the host. Furthermore, they contribute to the development and regulation of the immune system, promoting immune tolerance and defending against pathogenic microbes. The composition and balance of the microbiota are influenced by a variety of factors, including host genetics, diet, environmental exposures, and lifestyle choices [27, 28].
Distinct microbiomes have developed across various anatomical regions of the human body, each exhibiting unique characteristics and functions that collectively support overall health. The oral microbiome, first observed by Antonie van Leeuwenhoek in 1670 using his self-designed microscope, is the second-most diverse microbiome after the gut microbiome. Over 700 bacterial species have been identified within the oral cavity, which is subdivided into numerous niches, including saliva, the hard and soft palates, the tongue, lips, cheeks, and dental plaque biofilms [29, 30]. These distinct regions harbor specific microbial communities, which play vital roles in maintaining oral health, such as aiding in the digestion of food and protecting against pathogenic invaders. Disruption of the oral microbiome’s balance, a condition known as oral dysbiosis, is strongly associated with a range of oral diseases, including periodontitis and dental caries. The shift towards a pathogenic microbial profile often exacerbates inflammation, leading to tissue destruction and further compromising oral health [31, 32]. Recent research has highlighted the emerging links between oral dysbiosis and systemic diseases. The mechanisms behind these associations suggest that oral bacteria and their metabolites can enter the bloodstream through oral epithelial barriers or via direct interactions with the respiratory and digestive systems, thereby influencing systemic health [33,34,35]. Thus, the oral microbiome serves not only as a critical determinant of oral health but also as a potential modulator of broader health outcomes.
Historically, the respiratory tract was considered a sterile environment, with no microbial life present, a belief that endured for centuries. However, the advent of modern sequencing technologies, particularly 16S rRNA analysis, has revolutionized our understanding of the respiratory system. These advancements have demonstrated that the respiratory tract is not devoid of microorganisms, but rather supports a complex and diverse microbiome across both the upper and lower airways [36,37,38]. These respiratory microbiomes are shaped by a variety of factors, including environmental exposures, host genetics, and overall health status. Recent insights into the respiratory microbiota highlight its essential role in mucosal immunity and the regulation of inflammatory responses, which are key to maintaining respiratory health. Disruptions or imbalances in this microbiome—referred to as respiratory dysbiosis—have been linked to a range of chronic respiratory conditions, including asthma, COPD, and recurrent respiratory infections [39,40,41]. This growing body of evidence underscores the importance of the respiratory microbiota in both disease pathogenesis and the maintenance of pulmonary homeostasis, revealing its potential as a target for therapeutic interventions in respiratory diseases.
The gut microbiome is one of the most complex and densely populated microbial ecosystems in the human body, with trillions of microorganisms colonizing both the large and small intestines. This intricate community plays a central role in nutrient absorption, as well as the synthesis of essential metabolites such as short-chain fatty acids (SCFAs), which are crucial for maintaining intestinal health and regulating gut motility [42,43,44]. In addition to its metabolic functions, the gut microbiota exerts a profound influence on both local and systemic immune responses, shaping immune system development and regulating inflammatory pathways. Alterations in the composition of the gut microbiome, a condition known as gut dysbiosis, have been strongly associated with a range of diseases, including inflammatory bowel disease (IBD), metabolic syndrome, and various cancers. Disruptions in microbial diversity or the overgrowth of pathogenic species can compromise gut barrier function, facilitate systemic inflammation, and contribute to disease progression [45,46,47]. These findings underscore the pivotal role of the gut microbiome not only in intestinal health but also in the pathogenesis of a broad spectrum of systemic diseases, highlighting its potential as both a therapeutic target and a biomarker for disease detection.
Collectively, these interrelated microbial communities are essential for both localized functions and systemic homeostasis. Lifestyle factors, particularly smoking, have emerged as significant disruptors of these ecosystems. CS alters the composition and diversity of the microbiota across multiple body sites, impairing microbial balance and contributing to disease pathogenesis. Understanding the specific mechanisms by which smoking induces microbial dysbiosis across different anatomical sites is crucial, as this knowledge may uncover novel pathways for disease prevention, early detection, and personalized treatment. Moreover, such insights could inform the development of targeted therapeutic strategies aimed at restoring microbial balance, offering promising opportunities for improving health outcomes in individuals affected by smoking-related diseases.
Interplay between cigarette smoking and the oral microbiome
Building upon the foundational understanding of human microbial ecosystems, the oral cavity represents the first and most directly exposed site to cigarette smoke, making it a critical interface for host–microbe–toxin interactions. The unique ecological complexity of the oral microbiome renders it highly sensitive to chemical and thermal insults from tobacco products. CS and its alternatives—including electronic and smokeless forms—profoundly reshape microbial composition, functional capacity, and host immune responses. These alterations not only initiate local inflammatory and metabolic disturbances but also propagate systemic effects through microbial translocation and immune modulation. The following sections delineate how different tobacco products perturb the oral microbiota, contributing to disease onset, progression, and potential reversibility upon cessation.
Traditional cigarettes
Traditional cigarettes profoundly influence the oral microbiome (Fig. 2 and Table 1). CS reduced microbial α-diversity and reshaped bacterial composition. Smokers showed increased abundance of Moryella, Bulleidia, Moraxella, and nitrite-producing species such as Actinomyces and Veillonella, which enhanced oral acidity. Smoking also upregulated pathways related to amino acid and nucleotide sugar metabolism. Co-occurrence analysis revealed positive correlations among smoker-enriched taxa and negative correlations with depleted ones, indicating that CS disrupted microbial networks and metabolic balance [48]. Heavy smoking further modified the oral microbiota, enriching Veillonella dispar, Leptotrichia spp., and Prevotella pleuritidis, while nicotine dependence was linked to higher levels of Streptobacillus hongkongensis, Fusobacterium massiliense, and Prevotella bivia. Functional profiling showed increased activity in tricarballylate utilization, lactate racemization, and xanthosine metabolism pathways [49]. Shotgun metagenomic data confirmed that smoking induces salivary dysbiosis, with elevated Prevotella and Megasphaera and reduced Neisseria, Oribacterium, Capnocytophaga, and Porphyromonas. The enrichment of Prevotella, often linked to inflammation and oral cancer, underscored the pathogenic potential of smoking-altered communities [50]. CS also increased Streptococcus mutans, Veillonella tobetsuensis, and Veillonella dispar while decreasing Lactobacillus species. Notably, S. mutans strains from smokers displayed enhanced anthracene biodegradation, whereas Lactobacillus strains showed variable activity. Co-culture experiments revealed that bacterial interactions suppressed anthracene degradation, suggesting that smoking-induced dysbiosis altered both microbial composition and metabolic function [51]. Smoking further reduced oropharyngeal diversity and enriched periodontal pathogens such as Bacillus and Burkholderia. Unlike the nasopharyngeal microbiota, which remained relatively stable, the oropharyngeal microbiome exhibited marked smoking-related alterations, reflecting its higher sensitivity to CS exposure [52].
Both conventional and electronic cigarettes induce oral microbial dysbiosis through overlapping yet distinct mechanisms. Cigarette smoke, composed mainly of tar, nicotine, and carbon monoxide (CO), perturbs oral microbial homeostasis by reducing protective commensal bacteria, enriching pathogenic species, lowering oral pH, and disrupting metabolic processes such as xanthosine metabolism. In contrast, e-cigarette aerosols containing propylene glycol, nicotine, and flavoring agents promote microbial diversity shifts, enhance biofilm formation, and trigger immune dysregulation characterized by elevated pro-inflammatory cytokines (IL-6, IL-8, and IL-1β). These disturbances converge on oral microbial dysbiosis marked by ecological imbalance, metabolic disruption, immune dysfunction, and altered host gene expression. Collectively, these changes facilitate the onset and progression of oral diseases, including periodontitis, dental caries, recurrent aphthous stomatitis (RAS), oral malodor, and oral squamous cell carcinoma (OSCC). Created in https://BioRender.com.
Smoking-induced dysbiosis is a key driver of oral disease, particularly periodontitis. CS decreased Actinobacteria while increasing gram-negative anaerobes such as Fusobacterium and Campylobacter. Smokers showed lower abundance of Leptotrichia, Actinomyces, Corynebacterium, and Lautropia, indicating a microbial shift favoring periodontal pathogenesis [53]. Similarly, tobacco use altered the subgingival microbiome in chronic periodontitis, enriching Fusobacterium nucleatum, Neisseria sicca, and Veillonella dispar while depleting Prevotella species. These changes increase diversity yet promote dysbiosis, accelerating disease progression [54]. CS also elevated Actinomyces odontolyticus and reduced Streptococcus sanguinis, disrupting the balance between pathogenic and protective bacteria [55]. Moreover, CS altered microbial niches at multiple oral sites, enriching Treponema and Selenomonas in saliva and Dialister and Atopobium on the tongue. Genera such as Veillonella and Filifactor were found to correlate with cumulative exposure and were associated with periodontitis and halitosis, suggesting that the tongue might serve as a reservoir for pathogenic taxa in smokers [56].
Accumulating evidence indicates that smoking-related dysbiosis extends beyond periodontitis, contributing to caries, recurrent aphthous stomatitis (RAS), and oral squamous cell carcinoma (OSCC). Cigarette and alternative tobacco use markedly alter the supragingival microbiome: Streptococcus species dominated in smokers with low caries, while Lactobacillus species increased in those with high caries indices. Medwakh smokers showed enrichment of periodontopathogens in low-caries subjects and higher Klebsiella pneumoniae levels in high-caries subjects, highlighting microbiota-based differences in caries susceptibility [57]. Tobacco smoking also aggravated dysbiosis in RAS, reducing diversity and altering taxa composition. Smokers with RAS exhibited elevated Veillonella, Rothia, and Sneathia and reduced Bacteroidales and Bacteroides. These microbial changes correlated with smoking frequency and activated pathways related to respiration and pathogenicity, indicating that CS exacerbated RAS through microbial modulation [58]. During OSCC development, CS decreased microbial diversity and reduced Bacteroidetes, Proteobacteria, and Lactobacillus while promoting Staphylococcus proliferation. Integrative analyses showed associations between microbial composition and host gene expression, such as CD74 positively correlated with Lactobacillus and PPP1R3C negatively with Bacteroidota, suggesting that smoking-induced microbial shifted might interact with host transcriptional programs to promote OSCC progression [59].
Electronic cigarettes
Electronic cigarettes (ECs), developed as a safer alternative to traditional cigarettes, were initially hypothesized to reduce nicotine dependence and diminish adverse health effects due to the absence of tar and incomplete combustion byproducts. However, recent findings have highlighted that EC aerosols, containing nicotine, propylene glycol, and flavoring agents, disrupt the oral microbiome equilibrium while also promoting oral inflammatory responses, potentially exacerbating oral tissue damage. For instance, the use of electronic smoking devices altered dental microbiocenosis, reducing resident plaque microflora and increasing colonization by opportunistic pathogens such as Streptococcus pneumoniae and S. pyogenes. Heating tobacco systems and vape use shifted microbial profiles, with higher frequencies of S. aureus and Corynebacterium xerosis, potentially impacting oral health through transient pathogen colonization [60]. In addition, EC use altered oral microbiota, increasing Porphyromonas and Veillonella abundance and elevating IL-6 and IL-1β levels, suggesting heightened inflammatory responses. Exposure to EC aerosols enhanced susceptibility to infection, as shown by increased inflammatory responses in Porphyromonas gingivalis- and Fusobacterium nucleatum-challenged epithelial and malignant cell lines, highlighting a dysbiotic microbiome and immune dysregulation in EC users [61]. Remarkably, EC use led to pathogen overrepresentation, increased virulence, and a strong pro-inflammatory response in the oral cavity, comparable to severe periodontitis. The carbon-rich glycol/glycerol vehicle in ECs altered biofilm architecture within 24 h of exposure. A machine-learning classifier based on metagenomic signatures effectively identified EC users, including those who use ECs alone or alongside cigarettes, raising concerns about the safety of ECs and challenging harm reduction claims [62]. Similarly, the use of EC altered the subgingival microbiome, enriching Fusobacterium and Bacteroidales (G-2), and shared similarities with the microbiomes of conventional smokers and nonsmokers. Pathogenic taxa such as Treponema and Porphyromonas correlated with increased cytokine levels and periodontal inflammation, positioning the EC microbiome as a distinct state with unique oral health risks, closely resembling that of conventional smokers [63].
Importantly, EC-induced oral dysbiosis disrupts microbial homeostasis, fostering pathogenic proliferation and metabolic alterations that contribute to a spectrum of oral pathologies, including periodontitis and oral carcinogenesis. For instance, EC aerosol exposure disrupted oral microbial balance by inhibiting the growth of commensal Streptococcus species (S. sanguinis and S. gordonii), while enhancing biofilm formation and attachment of the opportunistic pathogen S. mutans. S. mutans also showed increased hydrophobicity and coaggregation abilities, potentially facilitating its colonization and contributing to oral health issues. These results indicated that EC use might promote pathogenic bacteria while suppressing beneficial commensals, potentially leading to oral diseases like periodontitis and dental caries [64]. Similarly, EC use modified the oral microbiome in periodontitis patients, enriching Filifactor, Treponema, and Fusobacterium, with similarities to the microbial shifts seen in cigarette smokers. Increased abundance of Porphyromonas gingivalis and Fusobacterium nucleatum was observed, alongside elevated pro-inflammatory cytokines, including IFN-γ and TNF-α, which correlated with genera such as Dialister and Selenomonas. These changes exacerbated oral microbiome dysbiosis and periodontal disease progression [65]. Likewise, EC use increased oral microbiome dysbiosis, with higher α-diversity and altered β-diversity compared to non-users. Significant changes in microbial taxa, such as Actinomyces, Rothia, Neisseria, and Enterococcus, in subgingival sites mediated the association between EC use and gingival inflammation. These microbial shifts, along with changes in 71 KEGG pathways, highlighted the potential for EC use to contribute to periodontal disease through its impact on the oral microbiome [66]. Remarkably, EC exposure enhanced S. aureus attachment and biofilm formation on oral epithelial cells, promoting oral colonization. This was coupled with a reduction in immune response, as indicated by decreased IL-8, IL-6, and IL-1β secretion, and impaired clearance of S. aureus. EC exposure also increased COX2 expression, suggesting modulation of the oral inflammatory response. These effects potentially facilitated the progression of periodontitis and oral preneoplasia by promoting S. aureus colonization and altering immune function [67]. In addition, EC exposure disrupted oral microbiome diversity and metabolite profiles, with flavored EC aerosols containing nicotine leading to increased bacterial α-diversity. Metabolomics analysis revealed significant changes in metabolites, particularly those linked to oral cancer progression. These findings highlight the adverse impact of EC use on oral health, altering both microbiome composition and metabolic pathways in a 3D organotypic model of human oral mucosa [68].
In addition to the dysbiosis of oral microbiota induced by exclusive EC use, studies have further explored the synergistic effects of combined EC consumption with various types of tobacco products on oral microbial communities and their associations with oral pathologies. For instance, nicotine pouch and EC use, like conventional smoking, were associated with the presence of periodontopathogenic bacteria (Porphyromonas gingivalis, Tannerella forsythia, Fusobacterium nucleatum) in saliva, while no such pathogens were detected in non-tobacco users. Taken together, the findings implied that nicotine pouches and ECs might alter the oral microbiome and contribute to periodontal disease risk, though further quantitative studies are needed to confirm these preliminary results [69]. Additionally, EC altered the oral microbiome, with EC users showing increased Veillonella abundance and distinct β-diversity compared to non-EC users. Dual use of ECs and conventional cigarettes further enhanced microbial α-diversity and was associated with pathogenic taxa, underscoring the microbiome’s sensitivity to combined smoking behaviors [70]. Interestingly, both traditional CS and EC use modified the oral microbiome, though with distinct effects. Traditional cigarette smokers showed increased abundance of Actinomyces and Prevotella, while EC users exhibited higher Veillonella and lower Porphyromonas and Peptostreptococcus levels. Smoking disrupted the balance of oral flora, promoting anaerobic bacteria associated with dental decay and bad breath. ECs, while similarly impacting the oral microbiome, had a different effect compared to traditional cigarettes, necessitating further investigation into their links to oral diseases [71].
Smokeless tobacco
Smokeless tobacco—composed of fermented plant materials, alkaline additives (e.g., sodium carbonate), and carcinogenic nitrosamines—differs fundamentally from combustible cigarettes and aerosolized ECs by delivering nicotine and toxins through direct mucosal contact rather than inhalation, a distinction that reshapes its biological interactions and uniquely perturbs oral microbial ecosystems through pH modulation, xenobiotic exposure, and epithelial barrier disruption. For instance, smokeless tobacco use induced oral mycobiome dysbiosis, with reduced diversity in users with oral lesions. The fungal genus Pichia, enriched in these users, correlated positively with Starmerella and Fusarium and negatively with Alternaria. Functional analysis revealed increased pathotrophic and saprotrophic fungal activities, suggesting disrupted fungal growth regulation. These alterations might contribute to oral carcinogenesis in smokeless tobacco users [72]. Similarly, smokeless tobacco aqueous extracts (STAEs) from various brands modulated the growth and viability of oral bacteria in a concentration-dependent manner, with some strains exhibiting inhibited growth and others enhanced growth. Snuff STAEs showed more toxicity to oral bacteria than snus. While tobacco-specific N-nitrosamines had minimal effects on bacterial growth and viability, STAEs disrupted the oral microbial balance by promoting certain bacterial strains and inhibiting others. These alterations in oral bacterial ecology might have implications for oral health [73]. Moreover, smokeless tobacco use significantly disrupted oral microbiota composition, with varying effects depending on dosage. Exposure to 250 mg of Grizzly snuff increased the abundance of Firmicutes, Streptococcus, Actinomyces, and other genera, while decreasing Bacteroidetes and Fusobacteria. Bacterial diversity was reduced at lower doses (2.5 mg) but increased at higher doses (250 mg). These findings highlight the impact of smokeless tobacco on oral microbial communities, indicating potential implications for oral health and disease development [74].
Smokeless tobacco use increased oral pathogenic bacterial diversity and promoted dysbiosis, particularly in users with oral premalignant lesions (OPL). Genera such as Prevotella, Fusobacterium, and Veillonella were enriched, alongside functional shifts in nitrogen, nucleotide, and energy metabolism pathways. The presence of HPV-16 and EBV further associated with OPL development, highlighting a cancer-promoting microbial profile in smokeless tobacco users [75]. In addition, smokeless tobacco altered the oral microbiome in OSCC patients, significantly enriching genera such as Staphylococcus, Fusobacterium, and Campylobacter, known for producing tobacco-specific nitrosamines. These microbial changes correlated with oncogenesis-related gene functions, highlighting the role of the oral bacteriome in oral carcinogenesis among smokeless tobacco users [76]. Similarly, smokeless tobacco consumption altered the oral microbiome, increasing inflammation-associated species and resembling the microbiome of OSCC patients. Streptococcus abundance distinguished healthy microbiomes from those of smokeless-tobacco users and OSCC sites. OSCC-associated microbiomes showed enrichment of Gram-negative genera such as Prevotella, Capnocytophaga, and Fusobacterium, linked to the lipopolysaccharide biosynthesis pathway, highlighting their potential as markers for oral cancer [77]. Moreover, Toombak, a traditional smokeless tobacco product widely consumed in Sudan, is composed of fermented tobacco leaves mixed with alkaline additives and was associated with elevated risks of OPLs and OSCC. Toombak use altered the oral microbiome, increasing Staphylococcaceae, Corynebacterium_1, and Cardiobacterium while reducing Prevotella, Lactobacillus, and Candida. Corynebacterium_1 was enriched in early cancer stages and oral cancer samples, suggesting a role in carcinogenesis. Genera such as Stenotrophomonas and Schlegelella dominated the oral cancer microbiome of Toombak users, potentially contributing to metastasis and poor prognosis, highlighting microbiome modulations as a risk factor for oral cancer progression [78]. Likewise, a cross-sectional investigation examining Shammah consumption—a culturally entrenched smokeless tobacco practice in Arabian communities—demonstrated distinct taxonomic shifts in tongue dorsum microbial communities. Shammah used altered the tongue microbiome, with significant shifts in species composition, particularly enriching species with high acetaldehyde-production potential. Notably, Rothia mucilaginosa, Streptococcus sp. oral taxon 66, and Actinomyces meyeri were more abundant in shammah users, while Oribacterium asaccharolyticum was more abundant in non-users. These microbiome changes might contribute to oral carcinogenesis, highlighting the need for further investigation into the role of smokeless tobacco in altering oral microbial communities [79].
Several recent studies have conducted comparative analyses to investigate the differential impacts of smokeless tobacco products versus conventional CS on oral microbiota composition in human populations. Notably, evidence from a 2024 longitudinal cohort revealed that tobacco use altered the oral microbiome, with cigarette and smokeless tobacco users exhibiting increased bacterial diversity, including a higher abundance of Firmicutes and a lower abundance of Proteobacteria compared to non-users. Non-users had a greater relative abundance of Actinomyces, Haemophilus, Neisseria, Rothia, and Veillonella. Tobacco users also exhibited shifts in species abundance over time and the presence of opportunistic pathogens such as Neisseria subflava and Porphyromonas endodontalis, highlighting tobacco’s impact on oral microbial composition [80]. Likewise, 16S rRNA gene sequencing was employed in a cross-sectional analysis of the oral microbiome across three cohorts: traditional cigarette smokers, smokeless tobacco users, and healthy controls, aiming to characterize tobacco-associated microbial signatures. Smokers and smokeless tobacco users exhibited distinct oral microbiome profiles compared to healthy controls, with higher microbial diversity and significant compositional differences. Smokers showed increased abundance of Fusobacterium spp., Saccharibacterium spp., and Shuttleworthia, while smokeless tobacco users had elevated levels of Fusobacterium, Catonella, and Fretibacterium. Functional pathways related to amino acid metabolism, including glutamate and aspartate biosynthesis, were enriched in both groups. These microbial and metabolic alterations highlighted the role of the oral microbiome in tobacco-related diseases, including oral cancer, and suggested its potential for diagnostic and therapeutic applications [81].
Collectively, these studies demonstrate that smokeless tobacco induces oral microbiome dysbiosis characterized by enrichment of pro-inflammatory taxa (e.g., Fusobacterium and Prevotella), depletion of symbiotic species (e.g., Lactobacillus and Neisseria), and activation of carcinogenesis-linked metabolic pathways (nitrogen metabolism and LPS biosynthesis), with dose-dependent effects and product-specific virulence. The convergence of acetaldehyde-producing fungi, oncoviral cofactors (HPV/EBV), and nitrosamine-enriched bacterial consortia establishes a polymicrobial carcinogenic milieu, necessitating functional metagenomic studies to disentangle microbial contributions to OSCC progression and targeted interventions to restore mucosal homeostasis.
Reversibility of smoking’s impact on the oral microbiome
While smoking-induced dysregulation of the oral microbiome drives oral inflammation and carcinogenesis, the biological feasibility of post-cessation recovery—and whether this recovery can decouple between microbial communities and long-term health risks—remains a critical gap in guiding precision prevention strategies. Evidence indicated that CS reduced microbial α-diversity in the buccal mucosa but had limited effects on microbial diversity and composition in other oral and nasal sites. The oral microbiota exhibited marked site-specific heterogeneity, potentially contributing to its resilience against smoking-induced environmental perturbations [82]. Furthermore, tobacco smoking profoundly altered oral microbial composition, enriching Bifidobacterium, Lactobacillus, and the phylum Actinobacteria while depleting Proteobacteria, with notable taxa-level changes. These effects were consistent across African-American and European-American groups and were not observed in former smokers, highlighting the reversible nature of smoking-induced microbiota disruptions following cessation [83]. In addition, CS altered the tongue microbiome, decreasing Neisseria and Capnocytophaga while increasing Streptococcus and Megasphaera. These changes were associated with altered metagenomic pathways, including nitrate reduction and the tricarboxylic acid cycle. Former smokers’ microbiomes resembled those of never-smokers, suggesting reversibility of smoking-induced dysbiosis [84]. Notably, CS significantly altered the salivary microbiota, increasing the abundance of genera such as Streptococcus, Prevotella, and Veillonella, while decreasing Neisseria. These microbial shifts suggested that the salivary microbiome might be restored after smoking cessation. Linear Discriminant Analysis Effect Size revealed a microbial signature that could classify smokers and nonsmokers based on genus abundance. Further proteomics and metabolomics studies are necessary to explore bacterial endotoxins, xenobiotic metabolism, and their impact on bacterial interactions in the salivary microbiome [85]. Similarly, cigarette smoke exposure significantly altered the oropharyngeal microbiota composition, reducing its diversity, with over 60 taxa diminished after 6 months of exposure. This dysbiosis was reversible 3 months after smoke cessation. Lung infection with Streptococcus pneumoniae exacerbated lung damage and prolonged microbiota alterations compared to control groups. The data implied that while smoke exposure induced microbiota disruption and emphysema, structural lung damage alone did not maintain the altered microbiota, indicating that microbial shifts might not directly contribute to emphysema progression [86].
Several studies specifically examined the biological feasibility of EC discontinuation. For instance, EC use was shown to induce significant changes in the oral microbiome, with increased α-diversity in saliva and shifts in β-diversity in buccal mucosa compared to nonsmokers. EC users exhibited higher levels of Veillonella and Haemophilus in saliva, and a trend toward increased Staphylococcus aureus colonization in nasal samples. These changes were partially reversible with reduced vaping. The findings highlighted vaping’s impact on oral microbial diversity and composition, suggesting potential implications for oral health [87]. Furthermore, EC and CS significantly altered oral microbiome diversity and composition, increasing Prevotellaceae and decreasing Neisseria compared to nonsmokers. Smoking groups shared dominant phyla, including Proteobacteria, Firmicutes, and Bacteroidetes, but nonsmokers exhibited higher Actinobacteria and Corynebacterium, linked to distinct functional profiles. Smoking cessation restored the oral microbiome towards nonsmoker profiles, highlighting reversible effects on microbial structure and function [88].
Dynamic interactions of cigarette smoke with the respiratory tract microbiota
Following its initial interaction within the oral cavity, inhaled cigarette smoke continues to exert profound effects along the respiratory tract, where it encounters another complex microbial ecosystem. The respiratory microbiota, extending from the nasopharynx to the alveoli, plays a central role in maintaining mucosal immunity and regulating inflammatory tone. Disruption of this delicate microbial balance by cigarette smoke leads to persistent immune activation, epithelial injury, and heightened susceptibility to infection and chronic airway disease. Understanding how smoking remodels the respiratory microbiome provides crucial insight into the mechanisms linking tobacco exposure to disorders such as COPD, asthma, and lung cancer.
The respiratory tract was long considered a sterile environment due to methodological constraints of traditional culture-based techniques. This paradigm shifted with the advent of high-throughput sequencing technologies, particularly 16S rRNA gene profiling and shotgun metagenomics, which unveiled a complex ecosystem harboring several hundred to over a thousand microbial species across the nasopharynx to alveoli [89]. Crucially, these culture-independent approaches enabled detection of fastidious anaerobes like Prevotella and Fusobacterium, whose dysregulation is now implicated in smoking-related pathologies. Leveraging these technological advances, recent multi-omics investigations have delineated smoking-induced remodeling of respiratory microbiota, with these dysbiotic changes further correlating to heightened risks of diverse respiratory pathologies (Table 2). For instance, CS significantly altered the lower respiratory tract microbiome, increasing the abundance of pathogenic bacteria such as Acinetobacter, Bacillus, and Staphylococcus, while decreasing beneficial taxa like Lactobacillaceae. Dysbiosis of Proteobacteria and Firmicutes phyla correlated with increased inflammation markers like IL-6 and C-reactive protein (CRP). Functional predictions showed disrupted microbial pathways, including amino acid transport and DNA repair, in smokers. This imbalance in the lung microbiome may disrupt immune homeostasis and contribute to inflammation, suggesting potential therapeutic targets such as probiotics (Fig. 3C) [16]. Moreover, CS disrupted microbiota, enriching Streptococcaceae in the oropharynx, with efficient transfer to germ-free mice. Upon influenza A virus infection, mice with cigarette smoke-associated microbiota exhibited greater weight loss and disease severity compared to controls. Microbiota changes induced by CS exposure, independent of structural lung damage, exacerbated influenza outcomes, highlighting the role of microbial dysbiosis in respiratory disease progression [90].
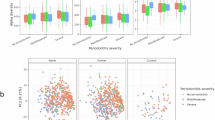
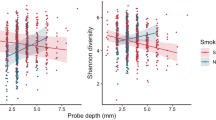
A In the nasal cavity, both cigarette smoke (CS) and e-cigarette (EC) exposure increased the abundance of Staphylococcus aureus, while Lactobacillus iners was enriched in CS users but depleted in EC users. B In the larynx, smoking reduced microbial diversity, marked by an increase in Streptococcus and a decrease in anaerobic or microaerophilic taxa such as Comamonadaceae (unclassified), Cloacibacterium, and Helicobacter. C In the lower respiratory tract, CS promoted the overgrowth of pathogenic taxa (Acinetobacter, Bacillus, Staphylococcus) and suppressed beneficial commensals, including Lactobacillaceae and Oceanospirillales. These changes disrupted microbial metabolic functions—such as amino acid transport, proline metabolism, and DNA repair—and were associated with elevated inflammatory mediators IL-6 and C-reactive protein (CRP). D In the lungs, Stenotrophomonas maltophilia overgrowth activated the interferon regulatory factor 1 (IRF1) and Z-DNA-binding protein 1 (ZBP1) signaling cascade, triggering PANoptosis and worsening alveolar damage characteristic of chronic obstructive pulmonary disease (COPD). Collectively, these findings illustrate how smoking-induced respiratory dysbiosis drives chronic inflammation, impairs tissue repair, and accelerates disease progression. Created in https://BioRender.com.
Substantial evidence has shown that CS-induced dysbiosis of the respiratory microbiota may drive COPD progression through disrupted microbial-host crosstalk and sustained airway inflammation. Notably, CS disrupted lung microbiota, promoting the expansion of Stenotrophomonas maltophilia in smoking-related COPD. S. maltophilia induced PANoptosis in alveolar epithelial cells via interferon regulatory factor 1 (IRF1)-mediated upregulation of Z-DNA Binding Protein 1 (ZBP1), impairing alveolar organoid formation. Targeting IRF1 mitigated S. maltophilia-induced lung injury, highlighting a link between microbial dysbiosis and emphysema progression in COPD (Fig. 3D) [91]. Furthermore, tobacco smoke induced lower airway dysbiosis in COPD, enriching oral commensals and promoting inflammatory pathways involving IL-17, IL-6, ERK/MAPK, and PI3K. These microbial and transcriptomic changes exacerbated inflammatory injury in early COPD, as corroborated in murine models, highlighting the interplay between smoking, airway microbiota, and disease pathogenesis [15].
Emerging studies suggest that CS-induced disruption of respiratory microbial communities may contribute to the pathogenesis of other respiratory diseases, including laryngitis, asthma and acute respiratory distress syndrome (ARDS). For instance, tobacco consumption reduced microbial diversity in the laryngeal microbiota, with notable shifts in the relative abundances of Streptococcus, Comamonadaceae, Cloacibacterium, and Helicobacter. Smokers exhibited less diversity compared to nonsmokers, while reflux status did not significantly impact microbial composition. The core laryngeal microbiota was dominated by Comamonadaceae, and increased Streptococcus abundance in benign vocal fold disease suggested its potential role in disease development. These findings highlight the impact of smoking on laryngeal microbiome structure and its possible contribution to chronic laryngitis (Fig. 3B) [92]. In addition, tobacco smoking in asthma patients increased bacterial diversity in the lungs compared to healthy nonsmokers, suggesting smoking influenced lung microbiota composition. However, smoking cessation did not significantly alter microbial diversity, indicating that while smoking exacerbated asthma symptoms and microbiome dysbiosis, cessation might not immediately restore microbial balance in the lungs [93]. Moreover, CS altered lung microbiota composition, with smokers showing enrichment of pathogens such as Streptococcus, Fusobacterium, Prevotella, Haemophilus, and Treponema. ARDS development after severe trauma was associated with microbial community shifts, including increased Enterobacteriaceae and specific taxa enriched in smokers, such as Prevotella and Fusobacterium. These results suggest smoking-related microbiota changes contribute to ARDS risk following trauma [94].
Comparative analyses of tobacco products have revealed divergent impacts on respiratory microbiota composition, with different tobacco products inducing distinct microbial alterations that may differentially modulate respiratory disease risks. EC and cigarette use induced significant dysbiosis in the nasal microbiome, with notable sex-dependent differences. Both EC users and smokers exhibited higher Staphylococcus aureus abundance, while Lactobacillus iners, a protective species, was less abundant in EC users. Dysbiosis also correlated with serum cotinine levels, indicating exposure to tobacco toxins. These results highlighted disrupted nasal immune responses and underscored the need for further research on the mechanisms by which ECs and smoking alter nasal immune homeostasis and microbiome composition, particularly considering sex as an important factor (Fig. 3A) [95]. Importantly, CS reduced bacterial abundance in the lung microbiome, with species such as Neisseria elongata and Haemophilus parainfluenzae significantly decreased. In contrast, EC use did not alter lung microbiota compared to never-smokers. Limited overlap between oral and lung microbiomes suggested the oral microbiome was not a reliable surrogate for smoking-related lung microbiome effects. These results underscored the distinct impact of cigarette smoke on microbial communities, potentially influencing disease risk [96]. Interestingly, a study investigating sub-chronic exposure to carbon nanotube (CNT) particles, cigarette smoke extract (CSE), and their combination on lung microbiota found that CSE altered lung microbiota, shifting from Proteobacteria to Bacteroidetes, while co-exposure with CNT resulted in mixed microbial effects, including increased Bacteroidetes and Tenericutes. CSE exposure also enriched pro-inflammatory oral genera such as Streptococcus and Aggregatibacter. These microbial changes correlated with disrupted lung mucosal homeostasis, highlighting the microbiome’s role in mediating smoking-induced respiratory toxicity [97].
Smoking-driven restructuring of gastrointestinal microbial communities
Beyond the respiratory tract, cigarette smoke exerts profound systemic effects that extend to the gastrointestinal ecosystem—the largest and most metabolically active microbial reservoir in the human body. The gut microbiota not only governs nutrient metabolism and immune homeostasis but also serves as a critical interface linking environmental exposures to host physiology. Continuous ingestion and inhalation of tobacco-derived toxins perturb this finely tuned ecosystem, leading to widespread alterations in microbial composition, metabolite profiles, and mucosal barrier integrity. These changes reverberate through interconnected axes such as the gut–lung and gut–liver pathways, amplifying inflammatory and metabolic dysfunctions. Understanding how cigarette smoke remodels gut microbial networks is therefore essential to delineate its contribution to systemic diseases and to identify novel microbiome-targeted strategies for prevention and therapy.
The human gastrointestinal tract harbors diverse microbial communities that maintain a delicate equilibrium through intricate cross-feeding networks and host–microbe metabolic interactions. These microbial communities, comprising over 1000 bacterial species alongside archaea, fungi, and viruses, collectively orchestrate essential physiological functions ranging from nutrient metabolism to immune modulation [98, 99]. Building on previous discoveries, recent research has revealed that exogenous factors—particularly cigarette smoke exposure—can fundamentally remodel this dynamically balanced architecture via nicotine-mediated epigenetic pathways (Table 3). For instance, tobacco smoking altered gut microbiota composition, affecting taxa such as Intestinimonas, Catenibacterium, and Ruminococcaceae. Smoking also influenced the abundance of specific gut microbes, creating a positive feedback loop involving Actinobacteria, which might link parental smoking to early smoking initiation in children. Additionally, neurotransmitter-associated metabolites like tryptophan and tyrosine likely mediated the gut microbiome’s influence on smoking behavior. These results illuminated the bidirectional relationship between smoking and gut dysbiosis, underscoring the impact of tobacco use on microbial balance and its potential role in smoking behaviors [100]. Moreover, CS altered both host gene expression and the gut microbiome, with 71 differential species and 324 differentially expressed genes identified between smokers and nonsmokers. Smoking influenced the gut microbiome through changes in heme metabolism, particularly affecting Bacteroides finegoldii and Lachnospiraceae bacterium 9_1_43BFAA. Bidirectional mediation analysis revealed that smoking modulated gut microbes via gene expression, with key metabolites like porphobilinogen and bilirubin linked to microbial changes. These findings provided new perspectives on the role of heme metabolism in mediating the effects of smoking on the gut microbiome [20].
Recent advances have shed light on how thirdhand smoke (THS) exposure disrupts infant gut microbiota development by altering microbial colonization patterns and metabolic cross-talk during critical early-life stages. Exposure to THS during postnatal development significantly altered gut microbiome composition in mice, with minimal effects observed during pubescent or adult exposure. Postnatal THS exposure increased degradation pathways related to glycolysis and pyruvate decarboxylation, while decreasing coenzyme A biosynthesis and pyrimidine deoxyribonucleoside salvage pathways. These findings suggested that the gut microbiome was particularly sensitive to early-life THS exposure, highlighting the long-term impact of environmental tobacco toxins on microbial diversity [101]. Likewise, THS exposure in neonatal ICU infants altered gut microbiome composition, with lower α-diversity observed in infants from smoking households or those with higher surface nicotine levels. Reduced Bifidobacterium abundance correlated with increased urine cotinine and household smoking, suggesting tobacco-related exposures negatively impacted infant gut microbiome development [102].
Smoking-driven gut microbiota destabilization exacerbates Crohn’s pathogenesis and activates colorectal oncogenic cascades
Recent findings have highlighted that cigarette smoking disrupts gut microbial ecosystems, weakens intestinal barrier integrity, and triggers dysbiosis-driven inflammation. These alterations contribute to Crohn’s disease and activate carcinogenic pathways that promote colorectal tumorigenesis. Chronic smoke exposure markedly reshaped the gut microbiome, increasing Lachnospiraceae sp. activity in the colon and elevating Muc2, Muc3, and Muc4 expression in the ileum and colon. It also altered immune mediators—upregulating Cxcl2 and Il-6 while suppressing Ifn-γ and Tgf-β—thereby disturbing mucosal homeostasis through concurrent shifts in microbiota, mucus composition, and cytokine signaling (Fig. 4A) [17]. Cigarette smoke condensate (CSC) aggravated inflammation and impaired Paneth cell function, reducing antimicrobial peptide secretion and bactericidal capacity. This led to fecal microbiota imbalance and increased susceptibility to bacterial injury. In IL-10(−/−) mice, CSC exposure induced severe enterocolitis, highlighting the role of smoke in disrupting intestinal equilibrium and promoting Crohn’s-like pathology (Fig. 4B) [103]. In experimental colitis, cigarette smoke modulated both the gut microbiome and colon transcriptome in a concentration-dependent manner. It altered dextran sodium sulfate–induced dysbiosis, affecting bacterial genera that could either resolve or sustain inflammation. Interestingly, epidemiological data indicated an inverse association between smoking and ulcerative colitis, suggesting complex tobacco–microbiota interactions that merit further study [104]. Beyond inflammation, smoke-induced dysbiosis facilitated colorectal cancer (CRC) progression. Tobacco exposure enriched Eggerthella lenta while depleting Parabacteroides distasonis and Lactobacillus spp., elevating bile acid metabolites such as taurodeoxycholic acid that activated MAPK/ERK signaling and promoted epithelial proliferation (Fig. 4C) [105]. Moreover, cigarette smoking increased the risk of type II colorectal neoplasms, with risk magnitude varying by gut enterotype. Smokers exhibited enrichment of carcinogenic Escherichia–Shigella and depletion of beneficial Lachnospiraceae and Ruminococcaceae. These shifts reinforced chronic inflammation and oncogenic signaling, linking smoke-related dysbiosis to colorectal tumorigenesis [106].
Cigarette smoke (CS) profoundly alters gut microbial composition and intestinal immune homeostasis, leading to chronic inflammation and tumorigenesis. A Chronic CS exposure increased the abundance of Lachnospiraceae species in the colon and upregulated mucus-associated genes (Muc2, Muc3, Muc4) and pro-inflammatory cytokines (Cxcl2, Il-6), while suppressing anti-inflammatory mediators such as interferon-gamma (Ifn-γ) and transforming growth factor-beta (Tgf-β). These changes disrupted mucosal barrier integrity and immune equilibrium across intestinal segments. B Exposure to cigarette smoke condensate (CSC) induced Paneth cell dysfunction in the ileum, resulting in granule abnormalities, reduced secretion of antimicrobial peptides, and decreased bactericidal capacity. This cascade led to fecal microbiota imbalance and promoted ileal inflammation, mimicking early pathogenic features of Crohn’s disease, a subtype of IBD. C CS-driven dysbiosis, characterized by an increase in Eggerthella lenta and depletion of Parabacteroides distasonis and Lactobacillus species, elevated taurodeoxycholic acid (TDCA) levels and activated the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway. This process compromised gut barrier integrity and promoted colorectal tumorigenesis, linking microbial alterations to carcinogenic signaling. Together, these findings illustrate how smoking-induced microbial and immune perturbations synergize to drive intestinal inflammation and malignant transformation. Created in https://BioRender.com.
Cigarette smoke disrupts gut–lung axis communication through microbial metabolite alterations driving pulmonary inflammation and cancer progression
Interestingly, accumulating investigations demonstrate that cigarette smoke distorts gut–lung axis communication by altering microbial metabolite profiles, creating a pro-inflammatory milieu that fuels pulmonary deterioration in COPD and promotes oncogenic niches in lung adenocarcinoma (LUAD). For instance, cigarette smoke exposure induced dysbiosis in both lung and intestinal microbiomes, with notable changes in bacterial diversity. Lung microbiome analysis revealed an increase in Actinobacteriota, while intestinal microbiome exhibited decreases in Patescibacteria, Campilobacterota, and others. These microbiome alterations correlated with lung function decline, including reduced forced vital capacity (FVC) and increased inflammation markers. Dysbiosis patterns observed in smoke-exposed mice were similar to those in severe COPD patients, suggesting a systemic impact of smoking on microbial balance and lung health (Fig. 5A) [21]. In addition, CS exposure altered gut microbiota composition in both wild-type and Nlrp6-deficient mice, with NLRP6 playing a critical role in controlling lung inflammation. Nlrp6-deficient mice exhibited impaired airway inflammation and neutrophil recruitment, while antibiotic treatment reduced CS-induced lung inflammation. Gut microbiota transferred from Nlrp6-deficient to wild-type mice attenuated lung inflammation, highlighting an Nlrp6-dependent gut-to-lung axis influencing pulmonary inflammation in response to smoking (Fig. 5B) [107]. Moreover, CS-induced COPD disrupted gut microbiota, reducing Akkermansia and Escherichia-Shigella, and activated the STAT3/NCOA4 pathway, promoting oxidative stress, inflammation, and ferritinophagy. Polyphyllin B restored microbial balance, inhibited STAT3/NCOA4 signaling, and ameliorated lung injury by enhancing ferritin and LC3 expression and mitigating mitochondrial damage, highlighting its therapeutic potential in smoking-related lung disease [108]. Additionally, tobacco smoke reduced intestinal microbial diversity and altered microbiota composition in mice, correlating with transcriptomic changes in the lung and ileum. Increased expression of genes such as MMP12 and SPP1 in the lung resembled COPD-like alterations, while intestinal gene changes (CD79B, PAX5) paralleled those in Crohn’s disease. These findings highlighted the interconnected impact of cigarette smoke on lung and gut inflammation, microbiota, and gene expression, suggesting shared mechanisms underlying COPD and intestinal disorders [109]. Likewise, tobacco smoking reduced SCFA production, with lower plasma SCFA concentrations observed in smokers, correlating with impaired lung function. Smoking altered gut microbiota and decreased fecal SCFAs, which could be modulated by dietary fiber or antibiotics. While SCFA modulation mitigated inflammation and alveolar destruction in a COPD mouse model, it had no effect in the elastase-induced model, suggesting SCFAs might influence COPD pathogenesis through their anti-inflammatory properties, offering potential therapeutic targets (Fig. 5C) [110]. Notably, tobacco carcinogen exposure led to significant changes in the gut and lung microbiomes, particularly in species like Odoribacter, Alistipes, Akkermansia, and Ruminococcus, which correlated with LUAD development and immunotherapeutic response. These changes were linked to decreased SCFAs such as propionic and butyric acid. Additionally, loss of Lcn2 expression disrupted microbiome composition, highlighting the role of microbial dysbiosis in tobacco-associated LUAD development. These results underscored the importance of microbiome dynamics in cancer progression and therapeutic responses [111].
Cigarette smoke (CS) exerts bidirectional effects on the gut and lung microbiota, disrupting their homeostasis and amplifying inflammatory signaling along the gut–lung axis. A CS exposure induced microbial dysbiosis in both the respiratory and intestinal tracts. In the lungs, the relative abundance of Actinobacteriota increased, whereas in the gut, Patescibacteria, Campilobacterota, Deferribacterota, and Actinobacteriota decreased. These alterations were accompanied by elevated pro-inflammatory mediators, including interleukins (ILs), interferon-gamma (IFN-γ), and 8-isoprostane, as well as physiological impairments such as reduced forced vital capacity (FVC), airway wall thickening, and emphysematous changes, paralleling microbial and pathological patterns observed in patients with chronic obstructive pulmonary disease (COPD). B The NLR family pyrin domain containing 6 (NLRP6) inflammasome served as a pivotal regulator of smoke-induced pulmonary inflammation. Nlrp6−/− mice displayed attenuated neutrophilic inflammation and reduced expression of chemokines CXCL1 and CXCL5. Fecal microbiota transplantation from Nlrp6−/− donors into wild-type (WT) recipients recapitulated this anti-inflammatory phenotype, underscoring the role of NLRP6-dependent gut microbiota in shaping airway inflammatory responses through the gut–lung axis. C CS decreased fecal short-chain fatty acid (SCFA) production by depleting SCFA-producing microbes. Replenishing SCFAs via dietary fiber supplementation mitigated inflammation and alveolar destruction in smoke-exposed mice, whereas antibiotic-induced microbiota depletion exacerbated pulmonary inflammation and tissue injury. These findings reveal that maintaining SCFA metabolism and gut microbial balance may represent a promising therapeutic strategy for ameliorating smoking-related COPD. Created in https://BioRender.com.
Tobacco smoke disrupts gut microbiota homeostasis driving metabolic dysfunction and multi-organ pathologies through inflammatory-metabolic axis dysregulation
Tobacco smoke exposure disrupts gut microbial homeostasis, triggering metabolic dysregulation and systemic pathologies, including weight loss, cardiovascular disease, diabetes mellitus, and hepatic dysfunction through microbiota-derived inflammatory mediators and metabolite imbalances. For instance, chronic cigarette smoke exposure altered the cecal microbiome, reducing microbial diversity and decreasing Alistipes abundance, a genus positively associated with body weight. Sex-specific differences in microbial composition were observed, with ovariectomy shifting the female microbiome to resemble that of males. These results suggested a link between smoke-induced microbiome changes and weight loss in smokers [112]. In addition, CS induced region-specific shifts in digestive tract microbiota, reducing beneficial genera such as Clostridium and Turicibacter while increasing harmful genera like Desulfovibrio and Bilophila. Functional predictions suggested impaired amino acid, lipid, and propionate metabolism alongside activated antioxidant pathways. These microbial alterations, coupled with hyperglycemia and reduced insulin and leptin levels, highlighted the role of smoking-induced gut dysbiosis in chronic disease risk [113]. Furthermore, tobacco smoking disrupted gut microbiota composition and hepatic metabolism, increasing cholesterol accumulation and altering bile acid homeostasis, particularly under high-fat diets. Changes in primary bile acid distribution and reduced CYP8B1 expression might contribute to smoking-induced insulin resistance and metabolic dysfunction. These findings highlighted the microbiome’s role in mediating smoking-related hepatic disorders [114]. Likewise, tobacco smoke exposure induced gut microbiota imbalances and liver injury, with significant changes in lipid metabolism-related gene expression in the liver. Salmonella correlated with the upregulation of lipid metabolism genes, while Ligilactobacillus showed opposite trends. These results underscored the coordinated regulation of lipid metabolism by gut microbiota and liver function, revealing key gut–liver interactions affected by smoking [115]. Notably, CS has been causally linked to metabolic diseases, with gut microbiota acting as a key mediator. Mendelian randomization analysis revealed that smoking influenced gut microbiota composition, particularly Paraprevotella clara, which significantly mediated the association between smoking and type 2 diabetes. The data highlighted the genetic and microbial interplay in smoking-induced metabolic disorders, underscoring the gut microbiome’s critical role in disease development [116]. Moreover, smoking cigarettes exacerbated gut microbiota dysbiosis in hypertensive individuals, reducing microbial α-diversity and shifting enterotypes toward Prevotella-dominant profiles. Smokers with hypertension showed reduced enrichment of beneficial taxa such as Phycisphaera and Clostridium asparagiforme, alongside altered microbial functions, highlighting the detrimental impact of smoking on gut health and its potential role in cardiovascular risk [117]. Importantly, CS altered intestinal microbiota, with Actinobacteria negatively correlating with pack-years and Cyanobacteria positively correlating with CO levels. Smoking cessation increased Bacteroidetes, decreased Firmicutes, and modestly raised α-diversity, which was inversely associated with heart rate, systolic blood pressure, and CRP. These outcomes suggested links between smoking, microbiota composition, and cardiovascular risk factors [118].
Diet-pharma-cessation-microbe axis mitigates tobacco-driven gut dysbiosis
Multimodal interventions encompassing dietary modulation, pharmacotherapy, smoking abstinence, probiotic supplementation, and microbial metabolite administration demonstrate efficacy in counteracting tobacco-induced gut dysbiosis and restoring microbial equilibrium. For instance, fermented black barley, rich in polyphenols and flavonoids, mitigated these effects by restoring microbial diversity, decreasing Lactobacillus, Turicibacter, and Bifidobacterium abundances, and increasing Oscillospira and Ruminococcus. It also alleviated smoking-induced metabolic disturbances, highlighting its potential in counteracting gut and systemic disruptions associated with smoking (Fig. 6A) [119]. Additionally, cigarette smoke exposure in a COPD mouse model altered the intestinal microbiome, with changes in diversity, composition, and metabolism, including lysine degradation and phenylalanine metabolism. Bufei Huoxue capsule (BFHX) treatment improved pulmonary function and reduced inflammation, while also modulating gut microbiota. The treatment dynamically regulated microbiome diversity and composition, highlighting a potential therapeutic mechanism for COPD through gut–lung interactions (Fig. 6B) [120]. Notably, cigarette smoke exposure induced significant shifts in the intestinal microbiome, notably increasing Akkermansiaceae abundance, which was reversed upon cessation. Switching to modified-risk tobacco products (MRTPs) like carbon-heated tobacco product 1.2 (CHTP 1.2) increased Lactobacillaceae abundance. These microbial changes suggested that CS altered gut microbiome composition and gene expression, with potential implications for gut function and disease pathogenesis. These findings highlighted the role of the microbiome in mediating smoking-related health risks and the potential for MRTP to modulate these effects (Fig. 6C) [121]. Likewise, CS reduced bacterial diversity in the duodenal mucosa-associated microbiota, increasing Streptococcus, Veillonella, and Rothia abundance while decreasing Prevotella and Neisseria. These microbiota changes persisted partially in former smokers, suggesting incomplete restoration post-cessation and potentially contributing to smoking-related gastrointestinal diseases [18]. By contrast, tobacco consumption significantly altered gut microbiota composition, with current smokers showing increased Bacteroidetes and decreased Firmicutes and Proteobacteria compared to never-smokers. β-diversity analysis revealed significant differences between current and never-smokers, as well as between former and current smokers. No significant differences were found between never and former smokers, suggesting that smoking cessation did not immediately revert microbiota composition to baseline [122]. Interestingly, cigarette smoke exposure depleted beneficial gut microbiota, including Bifidobacterium longum, and impaired SCFA production, particularly butyrate. Administration of B. longum, regardless of acetate production capacity, alleviated cigarette smoke-induced lung inflammation, reduced inflammatory cytokine and adhesion factor expression, and restored cecal butyrate levels. This highlighted the potential of B. longum probiotics in mitigating cigarette smoke-induced gut–lung axis dysregulation and inflammation (Fig. 6D) [123]. Furthermore, Euglena gracilis extracts counteracted tobacco carcinogen-induced lung tumorigenesis by modulating gut microbiota and increasing metabolites such as triethanolamine, salicylate, and SCFAs like acetate, propionate, and butyrate. These metabolites induced cell cycle arrest and apoptosis in lung carcinoma cells, linking gut microbiota alterations to anti-cancer activity against tobacco smoke-related carcinogenesis [124].
Cigarette smoke (CS) exposure perturbs the gut–lung axis, yet several interventional strategies have shown potential in restoring microbial balance and reducing inflammation. A Dietary supplementation with fermented black barley, rich in polyphenols and flavonoids, attenuated smoke-induced lung inflammation in ICR mice and helped re-establish gut microbial homeostasis. This intervention decreased the abundance of Lactobacillus, Turicibacter, and Bifidobacterium while enriching Oscillospira and Ruminococcus, suggesting a regulatory effect on the gut microbiome. B In a CS-exposed chronic obstructive pulmonary disease (COPD) mouse model, administration of Bufei Huoxue (BFHX) capsules improved pulmonary function, alleviated emphysema and lung inflammation, and restored gut microbial diversity, indicating that gut–lung axis modulation might underlie its therapeutic efficacy. C Smoking cessation reversed smoke-associated microbial alterations, notably reducing the relative abundance of Akkermansiaceae. Transition to modified-risk tobacco products (MRTPs), such as CHTP 1.2 aerosols, increased Lactobacillaceae levels, implying partial recovery of microbiota composition compared with conventional cigarette exposure. D Probiotic and microbial metabolite supplementation provided additional protective effects. Oral administration of Bifidobacterium longum suppressed lung inflammation, reduced inflammatory cytokine expression, and restored cecal butyrate levels. Similarly, Euglena gracilis extracts enhanced short-chain fatty acid (SCFA) production and inhibited tobacco carcinogen-induced lung tumorigenesis by promoting cell cycle arrest and apoptosis. Created in https://BioRender.com.
Challenges
Recent advances have illuminated the multifaceted ways in which cigarette smoke reconfigures microbial composition, functional capacity, and metabolic activity across body sites. Yet, the translation of these associations into mechanistic understanding remains hindered by methodological variability, inter-individual heterogeneity, and the intrinsic complexity of host–microbe–environment interactions. A significant challenge in studying the impact of smoking on the microbiome is the inherent variability in microbiome composition between individuals. This variability arises from multiple factors, including genetics, diet, environment, and lifestyle. Genetic differences can shape immune responses and microbial populations, while diet influences microbial diversity by providing nutrients that favor certain bacteria. Environmental exposures, along with lifestyle behaviors such as exercise, alcohol consumption, and medication use, also significantly affect microbiome composition [125,126,127]. This natural heterogeneity complicates efforts to isolate the specific effects of smoking, as it becomes difficult to discern whether observed changes in microbial composition are due to smoking itself or interactions between smoking and other factors. For example, smokers with different baseline microbiome compositions may exhibit distinct responses to smoking. To overcome this challenge, future studies should incorporate larger, demographically diverse cohorts that better represent population-level diversity. Employing statistical frameworks capable of adjusting for microbial covariates, such as host genetics, diet, and environmental factors, will enhance the robustness of findings. Additionally, integrating host genotyping and dietary profiling with microbiome data will refine stratification strategies and facilitate the development of individualized microbial risk signatures linked to tobacco exposure. Multi-omics approaches, combining genomics, transcriptomics, and metabolomics, could also provide a comprehensive understanding of the interactions between smoking and microbial communities [128, 129].
Sampling microbial communities presents another significant challenge, as it directly impacts the accuracy and reliability of study results. In gut microbiome studies, fecal samples are commonly used as proxies for intestinal microbial populations. However, substantial differences exist between the fecal microbiota and mucosal microbiota across various regions of the intestine. This discrepancy limits the representativeness of fecal samples, especially when studying localized conditions or disease states. Microbial communities in different sections of the gut, such as the small intestine and colon, vary in species composition and functional capacity, meaning that fecal samples may not fully reflect the diversity and dynamics of the intestinal microbiome [130,131,132]. The respiratory microbiome presents additional challenges due to its low microbial biomass, which makes it more susceptible to contamination during both sampling and sequencing. The relatively small number of microorganisms in the respiratory tract, compared to other body niches, requires highly sensitive methods for detecting and characterizing microbial species. Contamination from external sources or the upper airway can also interfere with analysis, leading to inaccurate data interpretation [133, 134]. To address these challenges, future research must focus on methodological innovations. Standardizing minimally invasive mucosal biopsies, spatially resolved metagenomics, and contamination controls—such as synthetic spike-in standards and dual-indexed barcoding—will enhance the reproducibility and accuracy of microbiome data [135,136,137]. Additionally, noninvasive yet site-specific sampling strategies should be developed to improve ecological fidelity and ensure that microbial communities are adequately represented [138, 139].
While animal models provide invaluable insights into microbial colonization and host–microbe interactions, they exhibit notable limitations when translating findings to humans. Murine studies, in particular, often employ smoke exposure protocols that differ significantly from human smoking behaviors, such as variations in smoke composition, exposure methods, and involuntary exposure patterns. These differences complicate the ability to assess smoking-induced microbiome alterations and associated disease mechanisms in humans [140, 141]. To enhance the translational relevance of animal studies, future research should emphasize the use of humanized gnotobiotic models, which are colonized with microbiota derived from well-characterized cohorts of smokers and nonsmokers. Combined with controlled environmental exposure systems—such as whole-body smoke chambers—and longitudinal tracking of host physiological and immunological responses, these models could offer a more accurate representation of real-world smoking conditions [142, 143]. Additionally, integrating advanced imaging techniques with microbiome analyses could provide deeper insights into the spatial distribution and interactions of microbial populations within different anatomical niches.
Another area that requires further investigation is the potential role of microbial metabolites in disease progression. It has been increasingly recognized that microbial dysbiosis, driven by smoking, can alter the production of key metabolites that influence host health. Understanding how microbial metabolites contribute to disease onset and progression is crucial for developing therapeutic interventions. To explore this, future studies should incorporate advanced metabolomics techniques to profile microbial metabolites produced in response to smoking. Animal models colonized with microbiota derived from smokers could help elucidate the direct contributions of these metabolites to disease development. Additionally, integrating bacterial, viral, and fungal analyses with metabolite profiling could uncover critical components that influence disease pathophysiology. Identifying and functionally characterizing these microbial metabolites may lead to novel therapeutic strategies aimed at modulating the microbiome to mitigate disease risks and improve smoking cessation outcomes [144, 145].
Finally, the integration of microbiome-based interventions offers a promising avenue for combating the adverse effects of smoking on health. Strategies such as probiotics, prebiotics, and dietary modifications, which aim to restore microbial balance, could be explored as adjunct therapies to smoking cessation programs. Moreover, microbiome-targeted therapies, including the use of antimicrobial peptides or specific bacteriophages, may provide more direct means of modulating microbial communities to reduce disease risk. Clinical trials investigating these interventions are needed to assess their efficacy in preventing or treating smoking-related diseases, such as periodontitis, COPD, and cancer.
Conclusions and perspectives
CS imposes widespread systemic consequences not only through its direct toxicological insults but also by reshaping microbial ecosystems that are fundamental to host homeostasis. Mounting evidence indicates that smoking-induced dysbiosis across the oral, respiratory, and gut microbiota perturbs immune–metabolic balance, thereby contributing to the onset and progression of inflammatory, metabolic, and neoplastic diseases. Although multi-omics technologies have delineated characteristic microbial signatures associated with tobacco exposure, key questions regarding causality, reversibility, and therapeutic applicability remain unresolved. Progress is hindered by the confounding effects of host genetics, diet, and environment, the heterogeneity of microbial sampling and sequencing methodologies, and the limited fidelity of rodent models in recapitulating human smoking behaviors. Future investigations should integrate longitudinal human cohorts with mechanistic animal studies colonized by smoker-derived microbiota, which may provide causal insight into how specific taxa or metabolites mediate disease trajectories. Moreover, dynamic multi-omics profiling that couples microbial composition with host immune and metabolic states will be pivotal to identifying keystone species and functional nodes amenable to therapeutic manipulation.
Beyond mechanistic understanding, delineating smoking-related microbial dysbiosis opens a new avenue for precision microbiome-based interventions. Strategies such as targeted probiotic supplementation, prebiotic and dietary modulation, engineered bacteriotherapies, or postbiotic metabolite administration hold promise in restoring microbial equilibrium and attenuating inflammation in smokers. Yet, the translation of these insights into clinical application remains challenging. Inter-individual variability in microbiome composition, transient colonization following microbial supplementation, and the lack of standardized endpoints to evaluate therapeutic efficacy collectively impede progress. Furthermore, the causal hierarchies linking microbial shifts to host pathology are still incompletely defined, complicating the design of rational microbiota-targeted interventions. Overcoming these barriers will require harmonized analytical frameworks, integrative computational models, and ethically guided clinical trials that align microbiome modulation with personalized disease risk stratification.
As the field evolves, coupling microbial ecology with systems-level host responses may transform our understanding of tobacco-related pathogenesis. This integrative perspective has the potential to advance microbiota-centered prevention and treatment strategies, ultimately complementing traditional smoking-cessation and public health efforts to alleviate the global burden of tobacco-induced disease.
Data availability
No datasets were generated or analysed during the current study.
References
Collaborators GBDT. Spatial, temporal, and demographic patterns in prevalence of smoking tobacco use and attributable disease burden in 204 countries and territories, 1990-2019: a systematic analysis from the Global Burden of Disease Study 2019. Lancet. 2021;397:2337–60.
Pirie K, Peto R, Reeves GK, Green J, Beral V. Million Women Study C. The 21st century hazards of smoking and benefits of stopping: a prospective study of one million women in the UK. Lancet. 2013;381:133–41.
Jha P. The hazards of smoking and the benefits of cessation: a critical summation of the epidemiological evidence in high-income countries. Elife. 2020;9:e49979.
Schmid D, Ricci C, Behrens G, Leitzmann MF. Does smoking influence the physical activity and lung cancer relation? A systematic review and meta-analysis. Eur Epidemiol. 2016;31:1173–90.
Stampfli MR, Anderson GP. How cigarette smoke skews immune responses to promote infection, lung disease and cancer. Nat Rev Immunol. 2009;9:377–84.
Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, et al. Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation. 2018;137:e67–e492.
Christenson SA, Smith BM, Bafadhel M, Putcha N. Chronic obstructive pulmonary disease. Lancet. 2022;399:2227–42.
Purdue MP, Silverman DT. Clearing the air: summarizing the smoking-related relative risks of bladder and kidney cancer. Eur Urol. 2016;70:467–8.
Gandini S, Botteri E, Iodice S, Boniol M, Lowenfels AB, Maisonneuve P, et al. Tobacco smoking and cancer: a meta-analysis. Int J Cancer. 2008;122:155–64.
Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6.
Brenchley JM, Serrano-Villar S. From dysbiosis to defense: harnessing the gut microbiome in HIV/SIV therapy. Microbiome. 2024;12:113.
Yoo JS, Oh SF. Unconventional immune cells in the gut mucosal barrier: regulation by symbiotic microbiota. Exp Mol Med. 2023;55:1905–12.
Jiang Y, Zhou X, Cheng L, Li M. The impact of smoking on subgingival microflora: from periodontal health to disease. Front Microbiol. 2020;11:66.
Tamashiro R, Strange L, Schnackenberg K, Santos J, Gadalla H, Zhao L, et al. Smoking-induced subgingival dysbiosis precedes clinical signs of periodontal disease. Sci Rep. 2023;13:3755.
Sulaiman I, Wu BG, Chung M, Isaacs B, Tsay JJ, Holub M, et al. Lower airway dysbiosis augments lung inflammatory injury in mild-to-moderate chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2023;208:1101–14.
Li KJ, Chen ZL, Huang Y, Zhang R, Luan XQ, Lei TT, et al. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir Res. 2019;20:272.
Allais L, Kerckhof FM, Verschuere S, Bracke KR, De Smet R, Laukens D, et al. Chronic cigarette smoke exposure induces microbial and inflammatory shifts and mucin changes in the murine gut. Environ Microbiol. 2016;18:1352–63.
Shanahan ER, Shah A, Koloski N, Walker MM, Talley NJ, Morrison M, et al. Influence of cigarette smoking on the human duodenal mucosa-associated microbiota. Microbiome. 2018;6:150.
Liu Y, Lu L, Yang H, Wu X, Luo X, Shen J, et al. Dysregulation of immunity by cigarette smoking promotes inflammation and cancer: a review. Environ Pollut. 2023;339:122730.
Li J, Yang Z, Yuan W, Bao Z, Li MD. Heme metabolism mediates the effects of smoking on gut microbiome. Nicotine Tob Res. 2024;26:742–51.
Laiman V, Chuang HC, Lo YC, Yuan TH, Chen YY, Heriyanto DS, et al. Cigarette smoke-induced dysbiosis: comparative analysis of lung and intestinal microbiomes in COPD mice and patients. Respir Res. 2024;25:204.
Campos M, Cickovski T, Fernandez M, Jaric M, Wanner A, Holt G, et al. Lower respiratory tract microbiome composition and community interactions in smokers. Access Microbiol. 2023;5:acmi000497.v3.
Cameron SJ, Lewis KE, Huws SA, Lin W, Hegarty MJ, Lewis PD, et al. Metagenomic sequencing of the chronic obstructive pulmonary disease upper bronchial tract microbiome reveals functional changes associated with disease severity. PLoS ONE. 2016;11:e0149095.
Ross FC, Patangia D, Grimaud G, Lavelle A, Dempsey EM, Ross RP, et al. The interplay between diet and the gut microbiome: implications for health and disease. Nat Rev Microbiol. 2024;22:671–86.
Kunath BJ, De Rudder C, Laczny CC, Letellier E, Wilmes P. The oral-gut microbiome axis in health and disease. Nat Rev Microbiol. 2024;22:791–805.
Xiao Y, Louwies T, Mars RAT, Kashyap PC. The human microbiome-a physiologic perspective. Compr Physiol. 2024;14:5491–519.
Donald K, Finlay BB. Early-life interactions between the microbiota and immune system: impact on immune system development and atopic disease. Nat Rev Immunol. 2023;23:735–48.
Shao T, Hsu R, Rafizadeh DL, Wang L, Bowlus CL, Kumar N, et al. The gut ecosystem and immune tolerance. J Autoimmun. 2023;141:103114.
Deo PN, Deshmukh R. Oral microbiome: Unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23:122–8.
Li X, Liu Y, Yang X, Li C, Song Z. The oral microbiota: community composition, influencing factors, pathogenesis, and interventions. Front Microbiol. 2022;13:895537.
Kozak M, Pawlik A. The role of the oral microbiome in the development of diseases. Int J Mol Sci. 2023;24:5231.
Di Stefano M, Santonocito S, Polizzi A, Mauceri R, Troiano G, Lo Giudice A, et al. A reciprocal link between oral, gut microbiota during periodontitis: the potential role of probiotics in reducing dysbiosis-induced inflammation. Int J Mol Sci. 2023;24:1084.
Kleinstein SE, Nelson KE, Freire M. Inflammatory networks linking oral microbiome with systemic health and disease. J Dent Res. 2020;99:1131–9.
Chhibber-Goel J, Singhal V, Bhowmik D, Vivek R, Parakh N, Bhargava B, et al. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. NPJ Biofilms Microbiomes. 2016;2:7.
O’Dwyer DN, Kim JS, Ma SF, Ranjan P, Das P, Lipinski JH, et al. Commensal oral microbiota, disease severity, and mortality in fibrotic lung disease. Am J Respir Crit Care Med. 2024;209:1101–10.
Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol. 2016;78:481–504.
Lucas SK, Yang R, Dunitz JM, Boyer HC, Hunter RC. 16S rRNA gene sequencing reveals site-specific signatures of the upper and lower airways of cystic fibrosis patients. J Cyst Fibros. 2018;17:204–12.
Hernandez-Leyva AJ, Rosen AL, Tomera CP, Lin EE, Akaho EH, Blatz AM, et al. Upper and lower airway microbiota across infancy and childhood. Pediatr Res. 2025;98:1449–59.
Opron K, Begley LA, Erb-Downward JR, Freeman C, Madapoosi S, Alexis NE, et al. Lung microbiota associations with clinical features of COPD in the SPIROMICS cohort. NPJ Biofilms Microbiomes. 2021;7:14.
Wang Z, Maschera B, Lea S, Kolsum U, Michalovich D, Van Horn S, et al. Airway host-microbiome interactions in chronic obstructive pulmonary disease. Respir Res. 2019;20:113.
Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. 2017;140:63–75.
Portincasa P, Bonfrate L, Vacca M, De Angelis M, Farella I, Lanza E, et al. Gut microbiota and short chain fatty acids: implications in glucose homeostasis. Int J Mol Sci. 2022;23:1105.
Zheng Z, Tang J, Hu Y, Zhang W. Role of gut microbiota-derived signals in the regulation of gastrointestinal motility. Front Med. 2022;9:961703.
Hamamah S, Amin A, Al-Kassir AL, Chuang J, Covasa M. Dietary fat modulation of gut microbiota and impact on regulatory pathways controlling food intake. Nutrients. 2023;15:3365.
White MT, Sears CL. The microbial landscape of colorectal cancer. Nat Rev Microbiol. 2024;22:240–54.
Hassan NE, El-Masry SA, El Shebini SM, Ahmed NH, Mohamed TF, Mostafa MI, et al. Gut dysbiosis is linked to metabolic syndrome in obese Egyptian women: potential treatment by probiotics and high fiber diets regimen. Sci Rep. 2024;14:5464.
Sharma B, Agriantonis G, Twelker K, Ebelle D, Kiernan S, Siddiqui M, et al. Gut microbiota serves as a crucial independent biomarker in inflammatory bowel disease (IBD). Int J Mol Sci. 2025;26:2503.
Jia YJ, Liao Y, He YQ, Zheng MQ, Tong XT, Xue WQ, et al. Association between oral microbiota and cigarette smoking in the Chinese population. Front Cell Infect Microbiol. 2021;11:658203.
Al Bataineh MT, Dash NR, Elkhazendar M, Alnusairat DMH, Darwish IMI, Al-Hajjaj MS, et al. Revealing oral microbiota composition and functionality associated with heavy cigarette smoking. J Transl Med. 2020;18:421.
Wirth R, Maróti G, Mihók R, Simon-Fiala D, Antal M, Pap B, et al. A case study of salivary microbiome in smokers and non-smokers in Hungary: analysis by shotgun metagenome sequencing. J Oral Microbiol. 2020;12:1773067.
Moussa HA, Wasfi R, Abdeltawab NF, Megahed SA. High counts and anthracene degradation ability of Streptococcus mutans and Veillonella parvula isolated from the oral cavity of cigarette smokers and non-smokers. Front Microbiol. 2021;12:661509.
Paulo AC, Lança J, Almeida ST, Hilty M, Sá-Leão R. The upper respiratory tract microbiota of healthy adults is affected by Streptococcus pneumoniae carriage, smoking habits, and contact with children. Microbiome. 2023;11:199.
Prince Y, Davison GM, Davids SFG, Erasmus RT, Kengne AP, Raghubeer S, et al. The effect of cigarette smoking on the oral microbiota in a South African population using subgingival plaque samples. Heliyon. 2024;10:e31559.
Moon JH, Lee JH, Lee JY. Subgingival microbiome in smokers and non-smokers in Korean chronic periodontitis patients. Mol Oral Microbiol. 2015;30:227–41.
Bašić K, Peroš K, Bošnjak Z, Šutej I. Subgingival microbiota profile in association with cigarette smoking in young adults: a cross-sectional study. Dent J. 2021;9:150.
Suzuki N, Nakano Y, Yoneda M, Hirofuji T, Hanioka T. The effects of cigarette smoking on the salivary and tongue microbiome. Clin Exp Dent Res. 2022;8:449–56.
Al-Marzooq F, Al Kawas S, Rahman B, Shearston JA, Saad H, Benzina D, et al. Supragingival microbiome alternations as a consequence of smoking different tobacco types and its relation to dental caries. Sci Rep. 2022;12:2861.
Wang X, Luo N, Mi Q, Kong W, Zhang W, Li X, et al. Influence of cigarette smoking on oral microbiota in patients with recurrent aphthous stomatitis. J Investig Med. 2022;70:805–13.
Liang D, Ma X, Zhong X, Zhou Y, Chen W, He X. Integration of host gene regulation and oral microbiome reveals the influences of smoking during the development of oral squamous cell carcinoma. Front Oncol. 2024;14:1409623.
Tishchenko OV, Kryvenko LS, Gargina VV. Influence of smoking heating up tobacco products and e-cigarettes on the microbiota of dental plaque. Pol Merkur Lekarski. 2022;50:16–20.
Pushalkar S, Paul B, Li Q, Yang J, Vasconcelos R, Makwana S, et al. Electronic cigarette aerosol modulates the oral microbiome and increases risk of infection. iScience. 2020;23:100884.
Ganesan SM, Dabdoub SM, Nagaraja HN, Scott ML, Pamulapati S, Berman ML, et al. Adverse effects of electronic cigarettes on the disease-naive oral microbiome. Sci Adv. 2020;6:eaaz0108.
Thomas SC, Xu F, Pushalkar S, Lin Z, Thakor N, Vardhan M, et al. Electronic cigarette use promotes a unique periodontal microbiome. mBio. 2022;13:e0007522.
Catala-Valentin A, Bernard JN, Caldwell M, Maxson J, Moore SD, Andl CD. E-cigarette aerosol exposure favors the growth and colonization of oral Streptococcus mutans compared to commensal streptococci. Microbiol Spectr. 2022;10:e0242121.
Xu F, Pushalkar S, Lin Z, Thomas SC, Persaud JK, Sierra MA, et al. Electronic cigarette use enriches periodontal pathogens. Mol Oral Microbiol. 2022;37:63–76.
Park B, Koh H, Patatanian M, Reyes-Caballero H, Zhao N, Meinert J, et al. The mediating roles of the oral microbiome in saliva and subgingival sites between e-cigarette smoking and gingival inflammation. BMC Microbiol. 2023;23:35.
Cátala-Valentín AR, Almeda J, Bernard JN, Cole AM, Cole AL, Moore SD, et al. E-cigarette aerosols promote oral S. aureus colonization by delaying an immune response and bacterial clearing. Cells. 2022;11:773.
Maan M, UJP, Mohamed DA, Jalaleddine N, Abuzayeda M, Khamis AH, et al. The effects of electronic cigarettes on oral microbiome and metabolome in 3D tissue-engineered models. Int Dent J. 2024;75:2239–52.
Miluna-Meldere S, Rostoka D, Broks R, Viksne K, Ciematnieks R, Skadins I, et al. The effects of nicotine pouches and E-cigarettes on oral microbes: a pilot study. Microorganisms. 2024;12:1514.
Yang I, Rodriguez J, Young Wright C, Hu YJ. Oral microbiome of electronic cigarette users: a cross-sectional exploration. Oral Dis. 2023;29:1875–84.
Liu J, Yue Q, Zhang S, Xu J, Jiang X, Su Q, et al. A pilot study on oral microbiome in electronic cigarettes consumers versus traditional cigarettes smokers. Folia Microbiol. 2024;70:147–158.
Sajid M, Sharma P, Srivastava S, Hariprasad R, Singh H, Bharadwaj M. Smokeless tobacco consumption induces dysbiosis of oral mycobiome: a pilot study. Appl Microbiol Biotechnol. 2022;106:5643–57.
Liu M, Jin J, Pan H, Feng J, Cerniglia CE, Yang M, et al. Effect of smokeless tobacco products on human oral bacteria growth and viability. Anaerobe. 2016;42:152–61.
Jin J, Guo L, VonTungeln L, Vanlandingham M, Cerniglia CE, Chen H. Smokeless tobacco impacts oral microbiota in a Syrian Golden hamster cheek pouch carcinogenesis model. Anaerobe. 2018;52:29–42.
Sajid M, Sharma P, Srivastava S, Hariprasad R, Singh H, Bharadwaj M. Alteration of oral bacteriome of smokeless tobacco users and their association with oral cancer. Appl Microbiol Biotechnol. 2023;107:4009–24.
Srivastava A, Mishra S, Garg PK, Dubey AK, Deo SVS, Verma D. Comparative and analytical characterization of the oral bacteriome of smokeless tobacco users with oral squamous cell carcinoma. Appl Microbiol Biotechnol. 2022;106:4115–28.
Saxena R, Prasoodanan PKV, Gupta SV, Gupta S, Waiker P, Samaiya A, et al. Assessing the effect of smokeless tobacco consumption on oral microbiome in healthy and oral cancer patients. Front Cell Infect Microbiol. 2022;12:841465.
Sami A, Elimairi I, Ryan CA, Stanton C, Patangia D, Ross RP. Altered oral microbiome in Sudanese Toombak smokeless tobacco users carries a newly emerging risk of squamous cell carcinoma development and progression. Sci Rep. 2023;13:6645.
Halboub E, Al-Ak’hali MS, Alamir AH, Homeida HE, Baraniya D, Chen T, et al. Tongue microbiome of smokeless tobacco users. BMC Microbiol. 2020;20:201.
Chattopadhyay S, Malayil L, Chopyk J, Smyth E, Kulkarni P, Raspanti G, et al. Oral microbiome dysbiosis among cigarette smokers and smokeless tobacco users compared to non-users. Sci Rep. 2024;14:10394.
Gopinath D, Wie CC, Banerjee M, Thangavelu L, Kumar RP, Nallaswamy D, et al. Compositional profile of mucosal bacteriome of smokers and smokeless tobacco users. Clin Oral Investig. 2022;26:1647–56.
Yu G, Phillips S, Gail MH, Goedert JJ, Humphrys MS, Ravel J, et al. The effect of cigarette smoking on the oral and nasal microbiota. Microbiome. 2017;5:3.
Yang Y, Zheng W, Cai QY, Shrubsole MJ, Pei Z, Brucker R, et al. Cigarette smoking and oral microbiota in low-income and African-American populations. J Epidemiol Community Health. 2019;73:1108–15.
Sato N, Kakuta M, Uchino E, Hasegawa T, Kojima R, Kobayashi W, et al. The relationship between cigarette smoking and the tongue microbiome in an East Asian population. J Oral Microbiol. 2020;12:1742527.
Al-Zyoud W, Hajjo R, Abu-Siniyeh A, Hajjaj S. Salivary microbiome and cigarette smoking: a first of its kind investigation in Jordan. Int J Environ Res Public Health. 2019;17:256.
Hilty M, Wüthrich TM, Godel A, Adelfio R, Aebi S, Burgener SS, et al. Chronic cigarette smoke exposure and pneumococcal infection induce oropharyngeal microbiota dysbiosis and contribute to long-lasting lung damage in mice. Microb Genom. 2020;6:mgen000485.
Chopyk J, Bojanowski CM, Shin J, Moshensky A, Fuentes AL, Bonde SS, et al. Compositional differences in the oral microbiome of E-cigarette users. Front Microbiol. 2021;12:599664.
Wang X, Mi Q, Yang J, Guan Y, Zeng W, Xiang H, et al. Effect of electronic cigarette and tobacco smoking on the human saliva microbial community. Braz J Microbiol. 2022;53:991–1000.
Ibironke O, McGuinness LR, Lu SE, Wang Y, Hussain S, Weisel CP, et al. Species-level evaluation of the human respiratory microbiome. Gigascience. 2020;9:giaa038.
Wüthrich T, de Brot S, Richina V, Mostacci N, Baumann Z, Leborgne NGF, et al. Cigarette smoke-induced disordered microbiota aggravates the severity of influenza A virus infection. mSystems. 2024;9:e0079024.
Xia H, Lin J, Wang Y, Yu J, Wang H, Cheng C, et al. Stenotrophomonas maltophilia contributes to smoking-related emphysema through IRF1-triggered PANoptosis of alveolar epithelial cells. Environ Pollut. 2024;349:123913.
Jetté ME, Dill-McFarland KA, Hanshew AS, Suen G, Thibeault SL. The human laryngeal microbiome: effects of cigarette smoke and reflux. Sci Rep. 2016;6:35882.
Munck C, Helby J, Westergaard CG, Porsbjerg C, Backer V, Hansen LH. Smoking cessation and the microbiome in induced sputum samples from cigarette smoking asthma patients. PLoS ONE. 2016;11:e0158622.
Panzer AR, Lynch SV, Langelier C, Christie JD, McCauley K, Nelson M, et al. Lung microbiota is related to smoking status and to development of acute respiratory distress syndrome in critically ill trauma patients. Am J Respir Crit Care Med. 2018;197:621–31.
Hickman E, Roca C, Zorn BT, Rebuli ME, Robinette C, Wolfgang MC, et al. E-cigarette use, cigarette smoking, and sex are associated with nasal microbiome dysbiosis. Nicotine Tob Res. 2024;27:114–24.
Ying KL, Brasky TM, Freudenheim JL, McElroy JP, Nickerson QA, Song MA, et al. Saliva and lung microbiome associations with electronic cigarette use and smoking. Cancer Prev Res. 2022;15:435–46.
Yadav B, Bhattacharya SS, Rosen L, Nagpal R, Yadav H, Yadav JS. Oro-respiratory dysbiosis and its modulatory effect on lung mucosal toxicity during exposure or co-exposure to carbon nanotubes and cigarette smoke. Nanomaterials. 2024;14:314.
Manor O, Dai CL, Kornilov SA, Smith B, Price ND, Lovejoy JC, et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun. 2020;11:5206.
Pepke ML, Hansen SB, Limborg MT. Unraveling host regulation of gut microbiota through the epigenome-microbiome axis. Trends Microbiol. 2024;32:1229–40.
Fan J, Zhou Y, Meng R, Tang J, Zhu J, Aldrich MC, et al. Cross-talks between gut microbiota and tobacco smoking: a two-sample Mendelian randomization study. BMC Med. 2023;21:163.
He L, Zhou YX, Zhang Y, Hang B, Chang H, Schick SF, et al. Thirdhand cigarette smoke leads to age-dependent and persistent alterations in the cecal microbiome of mice. Microbiologyopen. 2021;10:e1198.
Northrup TF, Stotts AL, Suchting R, Matt GE, Quintana PJE, Khan AM, et al. Thirdhand smoke associations with the gut microbiomes of infants admitted to a neonatal intensive care unit: an observational study. Environ Res. 2021;197:111180.
Berkowitz L, Pardo-Roa C, Salazar GA, Salazar-Echegarai F, Miranda JP, Ramírez G, et al. Mucosal exposure to cigarette components induces intestinal inflammation and alters antimicrobial response in mice. Front Immunol. 2019;10:2289.
Lo Sasso G, Phillips BW, Sewer A, Battey JND, Kondylis A, Talikka M, et al. The reduction of DSS-induced colitis severity in mice exposed to cigarette smoke is linked to immune modulation and microbial shifts. Sci Rep. 2020;10:3829.
Bai X, Wei H, Liu W, Coker OO, Gou H, Liu C, et al. Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. Gut. 2022;71:2439–50.
Cai JA, Zhang YZ, Yu ED, Ding WQ, Li ZS, Zhong L, et al. Association of cigarette smoking with risk of colorectal cancer subtypes classified by gut microbiota. Tob Induc Dis. 2023;21:99.
Nascimento M, Huot-Marchand S, Fanny M, Straube M, Le Bert M, Savigny F, et al. NLRP6 controls pulmonary inflammation from cigarette smoke in a gut microbiota-dependent manner. Front Immunol. 2023;14:1224383.
Wang Q, He Z, Zhu J, Hu M, Yang L, Yang H. Polyphyllin B inhibited STAT3/NCOA4 pathway and restored gut microbiota to ameliorate lung tissue injury in cigarette smoke-induced mice. BMC Biotechnol. 2024;24:13.
Wang L, Koelink PJ, Garssen J, Folkerts G, Henricks PAJ, Braber S. Gut microbiome and transcriptomic changes in cigarette smoke-exposed mice compared to COPD and CD patient datasets. Int J Mol Sci. 2024;25:4058.
Otake S, Chubachi S, Miyamoto J, Haneishi Y, Arai T, Iizuka H, et al. Impact of smoking on gut microbiota and short-chain fatty acids in human and mice: implications for COPD. Mucosal immunol. 2024;18:353–365.
Finnicum CT, Rahal Z, Hassane M, Treekitkarnmongkol W, Sinjab A, Morris R, et al. Pathogenesis of tobacco-associated lung adenocarcinoma is closely coupled with changes in the gut and lung microbiomes. Int J Mol Sci. 2022;23:10930.
Tam A, Filho FSL, Ra SW, Yang J, Leung JM, Churg A, et al. Effects of sex and chronic cigarette smoke exposure on the mouse cecal microbiome. PLoS ONE. 2020;15:e0230932.
Wang X, Ye P, Fang L, Ge S, Huang F, Polverini PJ, et al. Active smoking induces aberrations in digestive tract microbiota of rats. Front Cell Infect Microbiol. 2021;11:737204.
Yang Y, Yang C, Lei Z, Rong H, Yu S, Wu H, et al. Cigarette smoking exposure breaks the homeostasis of cholesterol and bile acid metabolism and induces gut microbiota dysbiosis in mice with different diets. Toxicology. 2021;450:152678.
Meng L, Xu M, Xing Y, Chen C, Jiang J, Xu X. Effects of cigarette smoke exposure on the gut microbiota and liver transcriptome in mice reveal gut-liver interactions. Int J Mol Sci. 2022;23:11008.
Zhang J, Hou L, Lei S, Li Y, Xu G. The causal relationship of cigarette smoking to metabolic disease risk and the possible mediating role of gut microbiota. Ecotoxicol Environ Saf. 2024;290:117522.
Wang P, Dong Y, Jiao J, Zuo K, Han C, Zhao L, et al. Cigarette smoking status alters dysbiotic gut microbes in hypertensive patients. J Clin Hypertens. 2021;23:1431–46.
Sublette MG, Cross TL, Korcarz CE, Hansen KM, Murga-Garrido SM, Hazen SL, et al. Effects of smoking and smoking cessation on the intestinal microbiota. J Clin Med. 2020;9:2963.
Zhong L, Qin L, Ding X, Ma L, Wang Y, Liu M, et al. The regulatory effect of fermented black barley on the gut microbiota and metabolic dysbiosis in mice exposed to cigarette smoke. Food Res Int. 2022;157:111465.
Li Y, Chen J, Xing Y, Wang J, Liang Q, Zeng J, et al. Bufei Huoxue capsule attenuates COPD-related inflammation and regulates intestinal microflora, metabolites. Front Pharm. 2024;15:1270661.
Battey JND, Szostak J, Phillips B, Teng C, Tung CK, Lim WT, et al. Impact of 6-month exposure to aerosols from potential modified risk tobacco products relative to cigarette smoke on the rodent gastrointestinal tract. Front Microbiol. 2021;12:587745.
Lee SH, Yun Y, Kim SJ, Lee EJ, Chang Y, Ryu S, et al. Association between cigarette smoking status and composition of gut microbiota: population-based cross-sectional study. J Clin Med. 2018;7:282.
Budden KF, Gellatly SL, Vaughan A, Amorim N, Horvat JC, Hansbro NG, et al. Probiotic Bifidobacterium longum subsp. longum protects against cigarette smoke-induced inflammation in mice. Int J Mol Sci. 2022;24:252.
Upreti D, Ishiguro S, Phillips M, Nakashima A, Suzuki K, Comer J, et al. Euglena gracilis extract protects from tobacco smoke carcinogen-induced lung cancer by altering gut microbiota metabolome. Integr Cancer Ther. 2023;22:15347354231195323.
Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–99.
Huda MN, Salvador AC, Barrington WT, Gacasan CA, D’Souza EM, Deus Ramirez L, et al. Gut microbiota and host genetics modulate the effect of diverse diet patterns on metabolic health. Front Nutr. 2022;9:896348.
Flynn JK, Ortiz AM, Herbert R, Brenchley JM. Host genetics and environment shape the composition of the gastrointestinal microbiome in nonhuman primates. Microbiol Spectr. 2023;11:e0213922.
Priya S, Burns MB, Ward T, Mars RAT, Adamowicz B, Lock EF, et al. Identification of shared and disease-specific host gene-microbiome associations across human diseases using multi-omic integration. Nat. Microbiol. 2022;7:780–95.
Zou S, Yang C, Zhang J, Zhong D, Meng M, Zhang L, et al. Multi-omic profiling reveals associations between the gut microbiome, host genome and transcriptome in patients with colorectal cancer. J Transl Med. 2024;22:175.
Carstens A, Roos A, Andreasson A, Magnuson A, Agreus L, Halfvarson J, et al. Differential clustering of fecal and mucosa-associated microbiota in ‘healthy’ individuals. J Dig Dis. 2018;19:745–52.
Yin XF, Ye T, Chen HL, Liu J, Mu XF, Li H, et al. The microbiome compositional and functional differences between rectal mucosa and feces. Microbiol Spectr. 2024;12:e0354923.
Vaga S, Lee S, Ji B, Andreasson A, Talley NJ, Agreus L, et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Sci Rep. 2020;10:14977.
Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87.
Biesbroek G, Sanders EA, Roeselers G, Wang X, Caspers MP, Trzcinski K, et al. Deep sequencing analyses of low density microbial communities: working at the boundary of accurate microbiota detection. PLoS ONE. 2012;7:e32942.
Hardwick SA, Chen WY, Wong T, Kanakamedala BS, Deveson IW, Ongley SE, et al. Synthetic microbe communities provide internal reference standards for metagenome sequencing and analysis. Nat Commun. 2018;9:3096.
Tourlousse DM, Ohashi A, Sekiguchi Y. Sample tracking in microbiome community profiling assays using synthetic 16S rRNA gene spike-in controls. Sci Rep. 2018;8:9095.
Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226.
Berlinberg AJ, Brar A, Stahly A, Gerich ME, Fennimore BP, Scott FI, et al. A novel approach toward less invasive multiomics gut analyses: a pilot study. Microbiol Spectr. 2022;10:e0244621.
Wang G, Menon S, Wilsack L, Rehak R, Lou L, Turbide C, et al. Spatially and temporally precise microbiome profiling in the small intestine using the SIMBA capsule with X-ray tracking. Front Microbiomes. 2024;3.
McGonigle P, Ruggeri B. Animal models of human disease: challenges in enabling translation. Biochem Pharmacol. 2014;87:162–71.
Van Norman GA. Limitations of animal studies for predicting toxicity in clinical trials: part 2: potential alternatives to the use of animals in preclinical trials. JACC Basic Transl Sci. 2020;5:387–97.
Park JC, Im SH. Of men in mice: the development and application of a humanized gnotobiotic mouse model for microbiome therapeutics. Exp Mol Med. 2020;52:1383–96.
Arrieta MC, Sadarangani M, Brown EM, Russell SL, Nimmo M, Dean J, et al. A humanized microbiota mouse model of ovalbumin-induced lung inflammation. Gut Microbes. 2016;7:342–52.
Chen Y, Chen Z, Liang L, Li J, Meng L, Yuan W, et al. Multi-kingdom gut microbiota dysbiosis is associated with the development of pulmonary arterial hypertension. EBioMedicine. 2025;115:105686.
Xiang B, Zhang Q, Wu H, Lin J, Xu Z, Zhang M, et al. Impact of mild COVID-19 history on oral-gut microbiota and serum metabolomics in adult patients with Crohn’s disease: potential beneficial effects. Biomedicines. 2024;12:2103.
Acknowledgements
The schematics were created with Biorender.com.
Funding
This work was supported by the National Natural Science Foundation of China (81901006, 82372905), the Young Top-notch Talent of Pearl River Talent Plan (0920220228), the Guangdong Provincial Science and Technology Project Foundation (2022A0505050038), Science and Technology Program of Guangzhou (No.2025A04J3464), and Science Research Cultivation Program of Stomatological Hospital, Southern Medical University (PY2020002, PY2022019).
Author information
Authors and Affiliations
Contributions
LC and SS contributed to conception and manuscript design. LC, SS, XZ, and ZZ drafted the manuscript. ZZ prepared the tables and figures. ZZ and XZ collected the references. LC, SS, XZ, and ZZ participated in the revision of the manuscript. All authors reviewed the manuscript and approved the final version of this manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, Z., Zhao, X., Sun, S. et al. Smoking-induced microbial dysbiosis: a key driver of systemic diseases and emerging therapeutic opportunities. Cell Death Discov. 12, 35 (2026). https://doi.org/10.1038/s41420-025-02914-x
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41420-025-02914-x