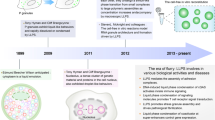
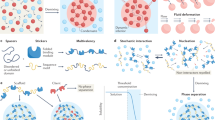
Abstract
Liquid‒liquid phase separation (LLPS) is a fundamental physical phenomenon in which a homogenous liquid spontaneously demixes into distinct liquid phases. A mounting body of evidence has shown that biomolecular LLPS is an essential biological event. In particular, highly condensed environments such as the nucleus are inevitably influenced by biomolecular LLPS, in which extremely long biopolymers, including genomic DNA and associated proteins/RNAs, are present. Given that almost half of the human genome is composed of repetitive elements and that various proteins interact with these sequences in diverse biological contexts, these regions clearly play substantial roles in regulating biomolecular LLPS. In this review, we summarize examples of biomolecular LLPS occurring in repetitive genomic elements. We also discuss how these intrinsic biophysical properties reflect cellular phenotypes by describing intermediate pathways and biomolecular complexes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hyman AA, Weber CA, Jülicher F. Liquid-Liquid Phase Separation in Biology. Annu Rev Cell Dev Biol. 2014;30:39–58. https://doi.org/10.1146/annurev-cellbio-100913-013325.
Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22:165–82. https://doi.org/10.1038/s41580-020-0272-6.
Caragine CM, Haley SC, Zidovska A. Nucleolar dynamics and interactions with nucleoplasm in living cells. eLife. 2019;8:e47533. https://doi.org/10.7554/eLife.47533.
Rippe K. Liquid–liquid phase separation in Chromatin. Cold Spring Harb Perspect Biol. 2022;14:a040683 https://doi.org/10.1101/cshperspect.a040683.
Frottin F, Schueder F, Tiwary S, Gupta R, Körner R, Schlichthaerle T, et al. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365:342–7. https://doi.org/10.1126/science.aaw9157.
Frank L, Rippe K. Repetitive RNAs as Regulators of chromatin-associated subcompartment formation by phase separation. Journal Mol Biol. 2020;432:4270–86. https://doi.org/10.1016/j.jmb.2020.04.015.
Park J, Kim J-J, Ryu J-K. Mechanism of phase condensation for chromosome architecture and function. Exp Mol Med. 2024;56:809–19. https://doi.org/10.1038/s12276-024-01226-x.
Li X, Liu C, Lei Z, Chen H, Wang L. Phase-separated chromatin compartments: Orchestrating gene expression through condensation. Cell Insight. 2024;3:100213. https://doi.org/10.1016/j.cellin.2024.100213.
Li X, An Z, Zhang W, Li F. Phase separation: direct and indirect driving force for high-order chromatin organization. Genes. 2023;14:499 https://doi.org/10.3390/genes14020499.
Hall AC, Ostrowski LA, Mekhail K. Phase separation as a melting pot for DNA repeats. Trends Genet. 2019;35:589–600. https://doi.org/10.1016/j.tig.2019.05.001.
Erdel F, Rippe K. Formation of Chromatin subcompartments by phase separation. Biophysical J. 2018;114:2262–70. https://doi.org/10.1016/j.bpj.2018.03.011.
Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. https://doi.org/10.1038/nature11247.
Palm W, De Lange T. How Shelterin Protects Mammalian Telomeres. Annu Rev Genet. 2008;42:301–34. https://doi.org/10.1146/annurev.genet.41.110306.130350.
Lim CJ, Cech TR. Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization. Nat Rev Mol Cell Biol. 2021;22:283–98. https://doi.org/10.1038/s41580-021-00328-y.
Cacchione S, Cenci G, Raffa GD. Silence at the End: How Drosophila Regulates Expression and Transposition of Telomeric Retroelements. Journal Mol Biol. 2020;432:4305–21. https://doi.org/10.1016/j.jmb.2020.06.004.
Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–40. https://doi.org/10.1038/nature05977.
Asamitsu S, Kawamoto Y, Hashiya F, Hashiya K, Yamamoto M, Kizaki S, et al. Sequence-specific DNA alkylation and transcriptional inhibition by long-chain hairpin pyrrole–imidazole polyamide–chlorambucil conjugates targeting CAG/CTG trinucleotide repeats. Bioorganic Med Chem. 2014;22:4646–57. https://doi.org/10.1016/j.bmc.2014.07.019.
Childs-Disney JL, Hoskins J, Rzuczek SG, Thornton CA, Disney MD. Rationally designed small molecules targeting the RNA that causes myotonic Dystrophy Type 1 are potently bioactive. ACS Chem Biol. 2012;7:856–62. https://doi.org/10.1021/cb200408a.
Nakatani K, Hagihara S, Goto Y, Kobori A, Hagihara M, Hayashi G, et al. Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats. Nat Chem Biol. 2005;1:39–43. https://doi.org/10.1038/nchembio708.
Asamitsu S, Yabuki Y, Ikenoshita S, Kawakubo K, Kawasaki M, Usuki S, et al. CGG repeat RNA G-quadruplexes interact with FMRpolyG to cause neuronal dysfunction in fragile X-related tremor/ataxia syndrome. Science Adv. 2021;7:eabd9440. https://doi.org/10.1126/sciadv.abd9440.
Hartley G, O’Neill RJ. Centromere repeats: hidden gems of the genome. Genes. 2019;10:223 https://doi.org/10.3390/genes10030223.
Wessler SR. Transposable elements and the evolution of eukaryotic genomes. Proc Natl Acad Sci. 2006;103:17600–1. https://doi.org/10.1073/pnas.0607612103.
Wells JN, Feschotte C. A field guide to eukaryotic transposable elements. Ann Rev Genet. 2020;54:539–61. https://doi.org/10.1146/annurev-genet-040620-022145.
Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18:71–86. https://doi.org/10.1038/nrg.2016.139.
Iwasaki YW, Shoji K, Nakagwa S, Miyoshi T, Tomari Y. (2025) Transposon–host arms race: a saga of genome evolution. Trends Genetics. https://doi.org/10.1016/j.tig.2025.01.009
Diehl AG, Ouyang N, Boyle AP. Transposable elements contribute to cell and species-specific chromatin looping and gene regulation in mammalian genomes. Nat Commun. 2020;11:1796. https://doi.org/10.1038/s41467-020-15520-5.
Choudhary MNK, Quaid K, Xing X, Schmidt H, Wang T. Widespread contribution of transposable elements to the rewiring of mammalian 3D genomes. Nat Commun. 2023;14:634. https://doi.org/10.1038/s41467-023-36364-9.
Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its biogenesis and functions. Ann Rev Biochem. 2015;84:405–33. https://doi.org/10.1146/annurev-biochem-060614-034258.
Brown RE, Freudenreich CH. Structure-forming repeats and their impact on genome stability. Current Opin Genet Dev. 2021;67:41–51. https://doi.org/10.1016/j.gde.2020.10.006.
Wang Y-L, Zhao W-W, Shi J, Wan X-B, Zheng J, Fan X-J. Liquid-liquid phase separation in DNA double-strand breaks repair. Cell Death Dis. 2023;14:1–10. https://doi.org/10.1038/s41419-023-06267-0.
Guo Y, Zhao S, Wang GG. Polycomb gene silencing mechanisms: PRC2 Chromatin Targeting, H3K27me3 “Readout”, and phase separation-based compaction. Trends Genet. 2021;37:547–65. https://doi.org/10.1016/j.tig.2020.12.006.
Asimi V, Sampath Kumar A, Niskanen H, Riemenschneider C, Hetzel S, Naderi J, et al. Hijacking of transcriptional condensates by endogenous retroviruses. Nat Genet. 2022;54:1238–47. https://doi.org/10.1038/s41588-022-01132-w.
Fueyo R, Judd J, Feschotte C, Wysocka J. Roles of transposable elements in the regulation of mammalian transcription. Nat Rev Mol Cell Biol. 2022;23:481–97. https://doi.org/10.1038/s41580-022-00457-y.
Lawson HA, Liang Y, Wang T. Transposable elements in mammalian chromatin organization. Nat Rev Genet. 2023;24:712–23. https://doi.org/10.1038/s41576-023-00609-6.
Bousios A, Nützmann H-W, Buck D, Michieletto D. Integrating transposable elements in the 3D genome. Mobile DNA. 2020;11:8. https://doi.org/10.1186/s13100-020-0202-3.
Jack A, Kim Y, Strom AR, Lee DSW, Williams B, Schaub JM, et al. Compartmentalization of telomeres through DNA-scaffolded phase separation. Dev Cell. 2022;57:277–290.e9. https://doi.org/10.1016/j.devcel.2021.12.017.
Kahn T, Savitsky M, Georgiev P. Attachment of HeT-A Sequences to Chromosomal Termini in Drosophila melanogaster may occur by different mechanisms. Mol Cell Biol. 2000;20:7634–42. https://doi.org/10.1128/MCB.20.20.7634-7642.2000.
George JA, DeBaryshe PG, Traverse KL, Celniker SE, Pardue M-L. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 2006;16:1231–40. https://doi.org/10.1101/gr.5348806.
McGurk MP, Dion-Côté A-M, Barbash DA. Rapid evolution at the Drosophila telomere: transposable element dynamics at an intrinsically unstable locus. Genetics. 2021;217:iyaa027. https://doi.org/10.1093/genetics/iyaa027.
Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin Protein 1 is involved in control of Telomere elongation in Drosophila melanogaster. Molecular Cell Biol. 2002;22:3204–18. https://doi.org/10.1128/MCB.22.9.3204-3218.2002.
Cenci G, Ciapponi L, Gatti M. The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma. 2005;114:135–45. https://doi.org/10.1007/s00412-005-0005-9.
Frydrychova RC, Mason JM, Archer TK. HP1 is distributed within distinct Chromatin domains at Drosophila Telomeres. Genetics. 2008;180:121–31. https://doi.org/10.1534/genetics.108.090647.
Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SIS. Role of Histone H3 Lysine 9 Methylation in epigenetic control of Heterochromatin assembly. Science. 2001;292:110–3. https://doi.org/10.1126/science.1060118.
Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, et al. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011;39:8703–11. https://doi.org/10.1093/nar/gkr552.
Teo RYW, Anand A, Sridhar V, Okamura K, Kai T. Heterochromatin protein 1a functions for piRNA biogenesis predominantly from pericentric and telomeric regions in Drosophila. Nat Commun. 2018;9:1735. https://doi.org/10.1038/s41467-018-03908-3.
Logsdon, Rozanski GA, Ryabov AN, PotapovaT F, Shepelev VA, Catacchio CR, et al. The variation and evolution of complete human centromeres. Nature. 2024;629:136–45. https://doi.org/10.1038/s41586-024-07278-3.
Roach RJ, Garavís M, González C, Jameson GB, Filichev VV, Hale TK. Heterochromatin protein 1α interacts with parallel RNA and DNA G-quadruplexes. Nucleic Acids Res. 2020;48:682–93. https://doi.org/10.1093/nar/gkz1138.
Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. https://doi.org/10.1002/1097-0134(20010101)42. 1<38::aid-prot50>3.0.co;2-3.
Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–40. https://doi.org/10.1038/nature22822.
Hiragami-Hamada K, Shinmyozu K, Hamada D, Tatsu Y, Uegaki K, Fujiwara S, et al. N-Terminal Phosphorylation of HP1α promotes its chromatin binding. Mol Cell Biol. 2011;31:1186–1200. https://doi.org/10.1128/MCB.01012-10.
Quivy J-P, Gérard A, Cook AJL, Roche D, Almouzni G. The HP1–p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–9. https://doi.org/10.1038/nsmb.1470.
Kiyomitsu T, Iwasaki O, Obuse C, Yanagida M. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J Cell Biol. 2010;188:791–807. https://doi.org/10.1083/jcb.200908096.
Huo X, Ji L, Zhang Y, Lv P, Cao X, Wang Q, et al. The Nuclear Matrix Protein SAFB cooperates with major satellite RNAs to stabilize Heterochromatin architecture partially through phase separation. Mol Cell. 2020;77:368–383.e7. https://doi.org/10.1016/j.molcel.2019.10.001.
Rivers C, Idris J, Scott H, Rogers M, Lee Y-B, Gaunt J, et al. iCLIP identifies novel roles for SAFB1 in regulating RNA processing and neuronal function. BMC Biol. 2015;13:111. https://doi.org/10.1186/s12915-015-0220-7.
Zenk F, Zhan Y, Kos P, Löser E, Atinbayeva N, Schächtle M, et al. HP1 drives de novo 3D genome reorganization in early Drosophila embryos. Nature. 2021;593:289–93. https://doi.org/10.1038/s41586-021-03460-z.
Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995;108:1419–31. https://doi.org/10.1242/jcs.108.4.1419.
Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–5. https://doi.org/10.1038/nature22989.
Tortora MMC, Brennan LD, Karpen G, Jost D. HP1-driven phase separation recapitulates the thermodynamics and kinetics of heterochromatin condensate formation. Proc Natl Acad Sci. 2023;120:e2211855120. https://doi.org/10.1073/pnas.2211855120.
Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–27. https://doi.org/10.1016/S0960-9822(01)00299-8.
Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, et al. The Small RNA Profile during Drosophila melanogaster Development. Dev Cell. 2003;5:337–50. https://doi.org/10.1016/S1534-5807(03)00228-4.
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. https://doi.org/10.1038/nature04916.
Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. https://doi.org/10.1038/nature04917.
Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–14. https://doi.org/10.1101/gad.1434406.
Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, et al. Characterization of the piRNA Complex from Rat Testes. Science. 2006;313:363–7. https://doi.org/10.1126/science.1130164.
Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating Loci as master regulators of Transposon Activity in Drosophila. Cell. 2007;128:1089–103. https://doi.org/10.1016/j.cell.2007.01.043.
Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and Endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. https://doi.org/10.1016/j.cell.2006.10.040.
Zhu C, Si X, Hou X, Xu P, Gao J, Tang Y, et al. (2023) Spatially clustered piRNA genes promote the transcription of piRNAs via condensate formation of the H3K27me3 reader UAD-2. bioRxiv 2023.12.10.571043
Hendriks IA, Vertegaal ACO. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol. 2016;17:581–95. https://doi.org/10.1038/nrm.2016.81.
Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43:1–9. https://doi.org/10.1016/j.pep.2005.03.016.
Thompson PJ, Dulberg V, Moon K-M, Foster LJ, Chen C, Karimi M, et al. hnRNP K coordinates transcriptional silencing by SETDB1 in embryonic stem cells. PLOS Genet. 2015;11:e1004933 https://doi.org/10.1371/journal.pgen.1004933.
Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–40. https://doi.org/10.1038/nature08674.
Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–31. https://doi.org/10.1038/nature08858.
Sripathy SP, Stevens J, Schultz DC. The KAP1 Corepressor functions to coordinate the assembly of De Novo HP1-demarcated microenvironments of heterochromatin required for KRAB Zinc Finger protein-mediated transcriptional repression. Molecular Cell Biol. 2006;26:8623–38. https://doi.org/10.1128/MCB.00487-06.
Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–9. https://doi.org/10.1038/nature08501.
Niki Y, Yamaguchi T, Mahowald AP. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci. 2006;103:16325–30. https://doi.org/10.1073/pnas.0607435103.
Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5’ end formation in Drosophila. Science. 2007;315:1587–90. https://doi.org/10.1126/science.1140494.
Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, et al. A piRNA pathway primed by individual transposons is linked to De Novo DNA methylation in mice. Mol Cell. 2008;31:785–99. https://doi.org/10.1016/j.molcel.2008.09.003.
Han BW, Wang W, Li C, Weng Z, Zamore PD. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015;348:817–21. https://doi.org/10.1126/science.aaa1264.
Mohn F, Handler D, Brennecke J. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348:812–7. https://doi.org/10.1126/science.aaa1039.
Murano K, Iwasaki YW, Ishizu H, Mashiko A, Shibuya A, Kondo S, et al. Nuclear RNA export factor variant initiates piRNA-guided co-transcriptional silencing. EMBO J. 2019;38:e102870. https://doi.org/10.15252/embj.2019102870.
Iwasaki YW, Murano K, Ishizu H, Shibuya A, Iyoda Y, Siomi MC, et al. Piwi Modulates Chromatin Accessibility by Regulating Multiple Factors Including Histone H1 to Repress Transposons. Mol Cell. 2016;63:408–19. https://doi.org/10.1016/j.molcel.2016.06.008.
Suyama R, Kai T piRNA processing within non-membrane structures is governed by constituent proteins and their functional motifs. FEBS J n/a: https://doi.org/10.1111/febs.17360
Gao J, Jing J, Shang G, Chen C, Duan M, Yu W, et al. TDRD1 phase separation drives intermitochondrial cement assembly to promote piRNA biogenesis and fertility. Dev Cell. 2024;59:2704–2718.e6. https://doi.org/10.1016/j.devcel.2024.06.017.
Hirakata S, Ishizu H, Fujita A, Tomoe Y, Siomi MC. Requirements for multivalent Yb body assembly in transposon silencing in Drosophila. EMBO Rep. 2019;20:e47708. https://doi.org/10.15252/embr.201947708.
Ohtani H, Iwasaki YW. Rewiring of chromatin state and gene expression by transposable elements. DGD. 2021;63:262–73. https://doi.org/10.1111/dgd.12735.
Schnabl J, Wang J, Hohmann U, Gehre M, Batki J, Andreev VI, et al. Molecular principles of Piwi-mediated cotranscriptional silencing through the dimeric SFiNX complex. Genes Dev. 2021;35:392–409. https://doi.org/10.1101/gad.347989.120.
Eastwood EL, Jara KA, Bornelöv S, Munafò M, Frantzis V, Kneuss E, et al. Dimerisation of the PICTS complex via LC8/Cut-up drives co-transcriptional transposon silencing in Drosophila. eLife. 2021;10:e65557. https://doi.org/10.7554/eLife.65557.
Ninova M, Chen Y-CA, Godneeva B, Rogers AK, Luo Y, Fejes Tóth K, et al. Su(var)2-10 and the SUMO Pathway Link piRNA-Guided Target Recognition to Chromatin Silencing. Mo Cell. 2020;77:556–570.e6. https://doi.org/10.1016/j.molcel.2019.11.012.
Andreev VI, Yu C, Wang J, Schnabl J, Tirian L, Gehre M, et al. Panoramix SUMOylation on chromatin connects the piRNA pathway to the cellular heterochromatin machinery. Nat Struct Mol Biol. 2022;29:130–42. https://doi.org/10.1038/s41594-022-00721-x.
Ninova M, Holmes H, Lomenick B, Fejes Tóth K, Aravin AA. Pervasive SUMOylation of heterochromatin and piRNA pathway proteins. Cell Genom. 2023;3:100329. https://doi.org/10.1016/j.xgen.2023.100329.
Iwasaki YW, Sriswasdi S, Kinugasa Y, Adachi J, Horikoshi Y, Shibuya A, et al. Piwi–piRNA complexes induce stepwise changes in nuclear architecture at target loci. EMBO J. 2021;40:e108345. https://doi.org/10.15252/embj.2021108345.
Zoch A, Auchynnikava T, Berrens RV, Kabayama Y, Schöpp T, Heep M, et al. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature. 2020;584:635–9. https://doi.org/10.1038/s41586-020-2557-5.
Dias Mirandela M, Zoch A, Leismann J, Webb S, Berrens RV, Valsakumar D, et al. Two-factor authentication underpins the precision of the piRNA pathway. Nature. 2024;634:979–85. https://doi.org/10.1038/s41586-024-07963-3.
Wang Y, Chen Y, Li M, Wang J, Jiang Y, Xie R, et al. Phase separation of SPIN1 through its IDR facilitates histone methylation readout and tumorigenesis. J Mol Cell Biol. 2024;16:mjae024. https://doi.org/10.1093/jmcb/mjae024.
Zamudio N, Bourc’his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap?. Heredity. 2010;105:92–104. https://doi.org/10.1038/hdy.2010.53.
Blackledge NP, Klose RJ. The molecular principles of gene regulation by Polycomb repressive complexes. Nat Rev Mol Cell Biol. 2021;22:815–33. https://doi.org/10.1038/s41580-021-00398-y.
Tatavosian R, Kent S, Brown K, Yao T, Duc HN, Huynh TN, et al. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. JBiol Chem. 2019;294:1451–63. https://doi.org/10.1074/jbc.RA118.006620.
Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, Marr SK, et al. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019;33:799–813. https://doi.org/10.1101/gad.326488.119.
Eeftens JM, Kapoor M, Michieletto D, Brangwynne CP. Polycomb condensates can promote epigenetic marks but are not required for sustained chromatin compaction. Nat Commun. 2021;12:5888. https://doi.org/10.1038/s41467-021-26147-5.
Niekamp S, Marr SK, Oei TA, Subramanian R, Kingston RE. Modularity of PRC1 composition and chromatin interaction define condensate properties. Mol Cell. 2024;84:1651–1666.e12. https://doi.org/10.1016/j.molcel.2024.03.001.
Saurin AJ, Shiels C, Williamson J, Satijn DPE, Otte AP, Sheer D, et al. The Human Polycomb Group Complex Associates with Pericentromeric Heterochromatin to Form a Novel Nuclear Domain. J Cell Biol. 1998;142:887–98. https://doi.org/10.1083/jcb.142.4.887.
Bracha D, Walls MT, Wei M-T, Zhu L, Kurian M, Avalos JL, et al. Mapping Local and Global Liquid Phase Behavior in Living Cells Using Photo-Oligomerizable Seeds. Cell. 2018;175:1467–1480.e13. https://doi.org/10.1016/j.cell.2018.10.048.
Hong Y, Bie L, Zhang T, Yan X, Jin G, Chen Z, et al. SAFB restricts contact domain boundaries associated with L1 chimeric transcription. Mol Cell. 2024;84:1637–1650.e10. https://doi.org/10.1016/j.molcel.2024.03.021.
Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell. 2017;168:159–171.e14. https://doi.org/10.1016/j.cell.2016.11.054.
Qian Z-G, Huang S-C, Xia X-X. Synthetic protein condensates for cellular and metabolic engineering. Nat Chem Biol. 2022;18:1330–40. https://doi.org/10.1038/s41589-022-01203-3.
Ninomiya K, Adachi S, Natsume T, Iwakiri J, Terai G, Asai K, et al. LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO J. 2020;39:e102729. https://doi.org/10.15252/embj.2019102729.
Ilık, İA, Glažar P, Tse K, Brändl B, Meierhofer D, et al. Autonomous transposons tune their sequences to ensure somatic suppression. Nature. 2024;626:1116–24. https://doi.org/10.1038/s41586-024-07081-0.
Tchasovnikarova IA, Timms RT, Matheson NJ, Wals K, Antrobus R, Göttgens B, et al. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348:1481–5. https://doi.org/10.1126/science.aaa7227.
Liu N, Lee CH, Swigut T, Grow E, Gu B, Bassik MC, et al. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature. 2018;553:228–32. https://doi.org/10.1038/nature25179.
Seczynska M, Bloor S, Cuesta SM, Lehner PJ. Genome surveillance by HUSH-mediated silencing of intronless mobile elements. Nature. 2022;601:440–5. https://doi.org/10.1038/s41586-021-04228-1.
Seczynska M, Lehner PJ. The sound of silence: mechanisms and implications of HUSH complex function. Trends Genet. 2023;39:251–67. https://doi.org/10.1016/j.tig.2022.12.005.
Prigozhin DM, Douse CH, Farleigh LE, Albecka A, Tchasovnikarova IA, Timms RT, et al. Periphilin self-association underpins epigenetic silencing by the HUSH complex. Nucleic Acids Res. 2020;48:10313–28. https://doi.org/10.1093/nar/gkaa785.
Lyu Y, Kim SJ, Humphrey ES, Nayak R, Guan Y, Liang Q, et al. Stem cell activity-coupled suppression of endogenous retrovirus governs adult tissue regeneration. Cell. 2024. https://doi.org/10.1016/j.cell.2024.10.007.
Han X, Xing L, Hong Y, Zhang X, Hao B, Lu JY, et al. Nuclear RNA homeostasis promotes systems-level coordination of cell fate and senescence. Cell Stem Cell. 2024;31:694–716.e11. https://doi.org/10.1016/j.stem.2024.03.015.
Shin Y, Chang Y-C, Lee DSW, Berry J, Sanders DW, Ronceray P, et al. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell. 2018;175:1481–1491.e13. https://doi.org/10.1016/j.cell.2018.10.057.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–21. https://doi.org/10.1126/science.1225829.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–23. https://doi.org/10.1126/science.1231143.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–6. https://doi.org/10.1126/science.1232033.
Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164:487–98. https://doi.org/10.1016/j.cell.2015.12.038.
Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Molecular Cell. 2017;68:808–820.e5. https://doi.org/10.1016/j.molcel.2017.10.015.
Padrón A, Iwasaki S, Ingolia NT. Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Molecular Cell. 2019;75:875–887.e5. https://doi.org/10.1016/j.molcel.2019.07.030.
Honda M, Oki S, Kimura R, Harada A, Maehara K, Tanaka K, et al. High-depth spatial transcriptome analysis by photo-isolation chemistry. Nat Commun. 2021;12:4416. https://doi.org/10.1038/s41467-021-24691-8.
Lee C, Quintana A, Suppanz I, Gomez-Auli A, Mittler G, Cissé II. Light-induced targeting enables proteomics on endogenous condensates. Cell. 2024;187:7079–7090.e17. https://doi.org/10.1016/j.cell.2024.09.040.
Acknowledgements
This work was supported by JSPS KAKENHI (Grant Numbers 23K13857 to S.A.; 24H02060, 22H02547 to Y.W.I.), RIKEN internal grants (Incentive Research Projects to S.A.), and JST FOREST (Grant Number JPMJFR224L to Y.W.I.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviews - special issue of Polymer Journal entitled “Current topics in liquid‒liquid phase separation”
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Asamitsu, S., Iwasaki, Y.W. Biomolecular liquid‒liquid phase separation associated with repetitive genomic elements. Polym J 57, 785–797 (2025). https://doi.org/10.1038/s41428-025-01036-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41428-025-01036-6