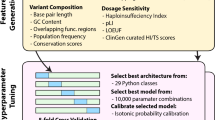
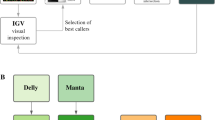
Abstract
Improved copy number variation (CNV) detection remains an area of heavy emphasis for algorithm development; however, both CNV curation and disease association approaches remain in its infancy. The current practice of focusing on candidate CNVs, where researchers study specific CNVs they believe to be pathological while discarding others, refrains from considering the full spectrum of CNVs in a hypothesis-free GWAS. To address this, we present a next-generation approach to CNV association by natively supporting the popular VCF specification for sequencing-derived variants as well as SNP array calls using a PennCNV format. The code is fast and efficient, allowing for the analysis of large (>100,000 sample) cohorts without dividing up the data on a compute cluster. The scripts are condensed into a single tool to promote simplicity and best practices. CNV curation pre and post-association is rigorously supported and emphasized to yield reliable results of highest quality. We benchmarked two large datasets, including the UK Biobank (n > 450,000) and CAG Biobank (n > 350,000) both of which are genotyped at >0.5 M probes, for our input files. ParseCNV has been actively supported and developed since 2008. ParseCNV2 presents a critical addition to formalizing CNV association for inclusion with SNP associations in GWAS Catalog. Clinical CNV prioritization, interactive quality control (QC), and adjustment for covariates are revolutionary new features of ParseCNV2 vs. ParseCNV. The software is freely available at: https://github.com/CAG-CNV/ParseCNV2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
• Project name: ParseCNV2
• Project (source code) home page: https://github.com/CAG-CNV/ParseCNV2
• Operating systems: Linux (32/64-bit), OS X (64-bit Intel), Windows (32/64-bit)
• Programming language: Perl, R, Bash
• Other requirements (when recompiling): none
• License: GNU General Public License version 3.0 (GPLv3)
• Any restrictions to use by non-academics: none
References
Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7.
Glessner JT, Li J, Hakonarson H. ParseCNV integrative copy number variation association software with quality tracking. Nucleic Acids Res. 2013;41:e64.
Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009;459:569–73.
Mace A, Tuke MA, Beckmann JS, Lin L, Jacquemont S, Weedon MN, et al. New quality measure for SNP array based CNV detection. Bioinformatics 2016;32:3298–305.
Glessner JT, Hou X, Zhong C, Zhang J, Khan M, Brand F, et al. DeepCNV: a deep learning approach for authenticating copy number variations. Brief Bioinform. 2021.
Kim JH, Hu HJ, Yim SH, Bae JS, Kim SY, Chung YJ. CNVRuler: a copy number variation-based case-control association analysis tool. Bioinformatics 2012;28:1790–2.
Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 2016;32:1423–6.
MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2014;42:D986–92.
Collins RL, Brand H, Karczewski KJ, Zhao X, Alfoldi J, Francioli LC, et al. A structural variation reference for medical and population genetics. Nature 2020;581:444–51.
Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015;526:75–81.
Werling DM, Brand H, An JY, Stone MR, Zhu L, Glessner JT, et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet. 2018;50:727–36.
Zhan X, Girirajan S, Zhao N, Wu MC, Ghosh D. A novel copy number variants kernel association test with application to autism spectrum disorders studies. Bioinformatics 2016;32:3603–10.
Alexander-Bloch A, Huguet G, Schultz LM, Huffnagle N, Jacquemont S, Seidlitz J, et al. Copy Number Variant Risk Scores Associated With Cognition, Psychopathology, and Brain Structure in Youths in the Philadelphia Neurodevelopmental Cohort. JAMA Psychiatry 2022;79:699–709.
Collins RL, Glessner JT, Porcu E, Lepamets M, Brandon R, Lauricella C, et al. A cross-disorder dosage sensitivity map of the human genome. Cell 2022;185:3041–55.e25.
Erikson GA, Deshpande N, Kesavan BG, Torkamani A. SG-ADVISER CNV: copy-number variant annotation and interpretation. Genet Med. 2015;17:714–8.
Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics 2018;34:3572–4.
Glessner JT, Bick AG, Ito K, Homsy J, Rodriguez-Murillo L, Fromer M, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115:884–96.
Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet. 2014;81:7 23 1–1.
Elia J, Glessner JT, Wang K, Takahashi N, Shtir CJ, Hadley D, et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet. 2011;44:78–84.
Aguirre M, Rivas MA, Priest J. Phenome-wide burden of copy-number variation in the UK Biobank. Am J Hum Genet. 2019;105:373–83.
Li YR, Glessner JT, Coe BP, Li J, Mohebnasab M, Chang X, et al. Rare copy number variants in over 100,000 European ancestry subjects reveal multiple disease associations. Nat Commun. 2020;11:1–9.
Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 2019;10:1784.
Greenside P, Zook J, Salit M, Cule M, Poplin R, DePristo M. CrowdVariant: a crowdsourcing approach to classify copy number variants. Pac Symp Biocomput. 2019;24:224–35.
Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 2017;33:2938–40.
Acknowledgements
We thank the study participants who allowed for the use of genotyping, sequencing, and disease phenotype data for this study, and to testers of the codes used in this study. Funding This work was supported in part by CHOP’s Endowed Chair in Genomic Research (Hakonarson), by 5U01HG011175-03 and U01-HG006830 (NHGRI-sponsored eMERGE Network), by a sponsored research agreement from Aevi Genomic Medicine Inc. (HH), Intellectual and Developmental Disabilities Research Center (IDDRC), Kids First Gabriella Miller Pediatric Research Program, and by an Institutional Development Award from Children’s Hospital of Philadelphia (HH).
Author information
Authors and Affiliations
Contributions
JTG conceived, designed, and implemented the code and wrote the paper. JL provided strategic guidance and ran other CNV association tools in benchmarking. YL provided and ran WES and WGS data CNV calls for validation of the ParseCNV2 algorithm and wrote those sections. MK compared ParseCNV2 with ParseCNV original version outputs to delineate reproducibility vs. new associations based on feature improvement. XC designed experiments and helped write the manuscript. PMAS contributed to data extraction. HH provided feedback on the report.
Corresponding author
Ethics declarations
Ethical approval
All subjects were recruited through IRB-approved protocols. Participants enrolled in various studies and completed a broad informed consent, including consent for prospective analyses of EHRs. Confidentiality is guarded to address issues of privacy and insurability. Each subject is assigned a study number upon recruitment, using complex algorithms to remove personal identification. Encrypted patient data is integrated into the lab’s custom phenotype browser, where it can be coupled with genotyping and sequencing data.
Competing interests
The authors declare no competing interests. Unrelated to this manuscript, we disclose that HH and CHOP own stock in Aevi Genomic Medicine.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Glessner, J.T., Li, J., Liu, Y. et al. ParseCNV2: efficient sequencing tool for copy number variation genome-wide association studies. Eur J Hum Genet 31, 304–312 (2023). https://doi.org/10.1038/s41431-022-01222-7
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-022-01222-7
This article is cited by
-
Genome-wide association studies for economically important traits in mink using copy number variation
Scientific Reports (2024)
-
Rare recurrent copy number variations in metabotropic glutamate receptor interacting genes in children with neurodevelopmental disorders
Journal of Neurodevelopmental Disorders (2023)
-
Genes=disease (?)
European Journal of Human Genetics (2023)
-
ParseCNV2: a versatile and integrated tool for copy number variation association studies
European Journal of Human Genetics (2023)