Abstract
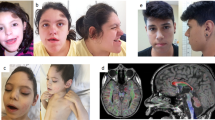
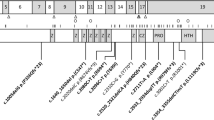
Haploinsufficiency of TRIP12 causes a neurodevelopmental disorder characterized by intellectual disability associated with epilepsy, autism spectrum disorder and dysmorphic features, also named Clark-Baraitser syndrome. Only a limited number of cases have been reported to date. We aimed to further delineate the TRIP12-associated phenotype and objectify characteristic facial traits through GestaltMatcher image analysis based on deep-learning algorithms in order to establish a TRIP12 gestalt. 38 individuals between 3 and 66 years (F = 20, M = 18) - 1 previously published and 37 novel individuals - were recruited through an ERN ITHACA call for collaboration. 35 TRIP12 variants were identified, including frameshift (n = 15) and nonsense (n = 6) variants, as well as missense (n = 5) and splice (n = 3) variants, intragenic deletions (n = 4) and two multigene deletions disrupting TRIP12. Though variable in severity, global developmental delay was noted in all individuals, with language deficit most pronounced. About half showed autistic features and susceptibility to obesity seemed inherent to this disorder. A more severe expression was noted in individuals with a missense variant. Facial analysis showed a clear gestalt including deep-set eyes with narrow palpebral fissures and fullness of the upper eyelids, downturned corners of the mouth and large, often low-set ears with prominent earlobes. We report the largest cohort to date of individuals with TRIP12 variants, further delineating the associated phenotype and introducing a facial gestalt. These findings will improve future counseling and patient guidance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data generated or analysed during this study are either included in this published article or available from the corresponding author on reasonable request. All variants were uploaded in the DECIPHER database (DECIPHER Patient ID 489845-489882) [13].
References
Clark RD, Baraitser M. A new X-linked mental retardation syndrome. Am J Med Genet. 1987;26:13–5.
Louie RJ, Friez MJ, Skinner C, Baraitser M, Clark RD, Schwartz CE, et al. Clark-Baraitser syndrome is associated with a nonsense alteration in the autosomal gene TRIP12. Am J Med Genet Part A U S. 2020;182:595–6.
Lelieveld SH, Reijnders MRF, Pfundt R, Yntema HG, Kamsteeg E-J, de Vries P, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–6.
O’Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014;515:216–21.
Bramswig NC, Lüdecke H-J, Pettersson M, Albrecht B, Bernier RA, Cremer K, et al. Identification of new TRIP12 variants and detailed clinical evaluation of individuals with non-syndromic intellectual disability with or without autism. Hum Genet. 2017;136:179–92.
Zhang J, Gambin T, Yuan B, Szafranski P, Rosenfeld JA, Al Balwi M, et al. Haploinsufficiency of the E3 ubiquitin-protein ligase gene TRIP12 causes intellectual disability with or without autism spectrum disorders, speech delay, and dysmorphic features. Hum Genet. 2017;136:377–86.
Donoghue T, Garrity L, Ziolkowski A, McPhillips M, Buckman M, Goel H. Novel de novo TRIP12 mutation reveals variable phenotypic presentation while emphasizing core features of TRIP12 variations. Am J Med Genet Part A U S. 2020;182:1801–6.
George AJ, Hoffiz YC, Charles AJ, Zhu Y, Mabb AM. A comprehensive atlas of E3 ubiquitin ligase mutations in neurological disorders. Front Genet. 2018;9:29.
Brunet M, Vargas C, Larrieu D, Torrisani J, Dufresne M. E3 ubiquitin ligase TRIP12: regulation, structure, and physiopathological functions. Int J Mol Sci. 2020;21:8515.
Hsieh T-C, Bar-Haim A, Moosa S, Ehmke N, Gripp KW, Pantel JT, et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat Genet. 2022;54:349–57.
Zhang Z, Song Y.Qi H, Age progression/regression by conditional adversarial autoencoder. IEEE Conf Comput Vis Pattern Recognit, CVPR 2017. 2017;4352–60.
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–33.
Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–74.
Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94.
Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–23.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–24.
Laan L Van Der, Rooney K, Alders M, Relator R, Mcconkey H, Kerkhof J, et al. Episignature mapping of TRIP12 provides functional insight into Clark–Baraitser syndrome. Int J Mol Sci. 2022;23:13664.
Styne DM, Arslanian SA, Connor EL, Farooqi IS, Murad MH, Silverstein JH, et al. Pediatric obesity-assessment, treatment, and prevention: an endocrine society clinical practice guideline. J Clin Endocrinol Metab. 2017;102:709–57.
Kajiro M, Tsuchiya M, Kawabe Y-I, Furumai R, Iwasaki N, Hayashi Y, et al. The E3 ubiquitin ligase activity of Trip12 is essential for mouse embryogenesis. PLoS One. 2011;6:e25871.
Acknowledgements
The authors would like to thank the patients and caregivers for their participation in this study. We are grateful to Caterina Lo Rizzo and Francesca Ariani for their contribution.
Funding
HVE is supported by a Senior Clinical Investigator fellowship of the Fonds voor Wetenschappelijk Onderzoek (FWO) Flanders. Almost all authors are members of the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability ERN-ITHACA [EU Framework Partnership Agreement ID: 3HP-HP-FPA ERN-01-2016/739516]. K.Õ is supported by grant from the Estonian Research Council (grant PRG471). The Broad Center for Mendelian Genomics (UM1 HG008900) is funded by the National Human Genome Research Institute with supplemental funding provided by the National Heart, Lung, and Blood Institute under the Trans-Omics for Precision Medicine (TOPMed) program and the National Eye Institute.
Author information
Authors and Affiliations
Contributions
All authors were involved in the collection of clinical information, the genetic analyses/data interpretation and/or the preparation of the manuscript and all approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was conducted according to the UZ Leuven ethical commission guidelines. Appropriate informed consent was obtained for each affected individual, in accordance with the respective local ethical committee. Written informed consent for the publication of pictures was obtained for 21 individuals.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Aerden, M., Denommé-Pichon, AS., Bonneau, D. et al. The neurodevelopmental and facial phenotype in individuals with a TRIP12 variant. Eur J Hum Genet 31, 461–468 (2023). https://doi.org/10.1038/s41431-023-01307-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-023-01307-x
This article is cited by
-
Novel TRIP12 variants in two Lebanese patients with neurodevelopmental delay
BMC Medical Genomics (2025)
-
The Role of a Novel TRIP12 Mutation in Intellectual Disability: A Molecular and Clinical Investigation in Multiplex Family
Journal of Molecular Neuroscience (2025)
-
Engineered targeting OIP5 sensitizes bladder cancer to chemotherapy resistance via TRIP12-PPP1CB-YBX1 axis
Oncogene (2024)
-
A Comprehensive Review of Syndromic Forms of Obesity: Genetic Etiology, Clinical Features and Molecular Diagnosis
Current Obesity Reports (2024)
-
April, again
European Journal of Human Genetics (2023)