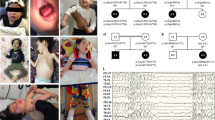
Abstract
Syndromes associating both eyeball and periocular developmental anomalies, combining iris chorioretinal (ocular) coloboma and ptosis, are described in very rare clinical entities such as Baraitser-Winter cerebrofrontofacial syndrome (BWCFF). We report on six individuals from 3 unrelated families presenting with autosomal dominant eye malformations, including ocular coloboma, ptosis and craniofacial features suggesting BWCFF. However, no neurodevelopmental disorders (NDD) as usually observed in this syndrome were detected. Exome sequencing (ES) or genome sequencing (GS) was performed and allowed the identification of 3 novel heterozygous variants in the MYH10 gene, encoding the non-muscle myosin heavy chain II B. These 3 likely causative variants occur in the MYH10 tail domain required for myosin filament assembly. The MYH10 protein is mislocalized leading to abnormal actin networks in the patients’ fibroblasts compared to controls. MYH10 dysfunction leads to delayed development of the eye, as well as a muscular phenotype in the zebrafish model. Heterozygous variants in MYH10 have been recently reported to be associated with an autosomal dominant NDD with other congenital anomalies, but no patients were reported with the association of ocular coloboma and ptosis as main features. Herein, we report other MYH10 variants which cause mainly an ophthalmic phenotype without NDD expanding the phenotype associated with MYH10 and representing a differential diagnosis with BWCFF. The reason for the genotype-phenotype variability with either prominent NDD or prominent ocular features will require further investigations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data generated or analysed during this study are included in the published article and the corresponding supplemental data. The raw sequencing data generated in the course of this study are not publicly available due to the protocol and the corresponding consent used that did not include such information. The variants identified have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) using the following accessions numbers SCV000898494, SCV000898495, SCV005043360 and SCV005043361.
References
Plaisancié J, Ceroni F, Holt R, Seco CZ, Calvas P, Chassaing N, et al. Genetics of anophthalmia and microphthalmia. Part 1: non-syndromic anophthalmia/microphthalmia. Hum Genet. 2019;138:799–830. Feb 14 [cited 2019 Sep 26]Available from: https://link.springer.com/article/10.1007/s00439-019-01977-y.
Fitzpatrick DR, van Heyningen V. Developmental eye disorders. Curr Opin Genet Dev. 2005;15:348–53.
Holtz AM, VanCoillie R, Vansickle EA, Carere DA, Withrow K, Torti E, et al. Heterozygous variants in MYH10 associated with neurodevelopmental disorders and congenital anomalies with evidence for primary cilia-dependent defects in Hedgehog signaling. Genet Med. 2022;24:2065–78.
Ma X, Kawamoto S, Hara Y, Adelstein RS. A point mutation in the motor domain of nonmuscle myosin II-B impairs migration of distinct groups of neurons. Mol Biol Cell. 2004;15:2568–79.
Ma X, Kawamoto S, Uribe J, Adelstein RS. Function of the neuron-specific alternatively spliced isoforms of nonmuscle myosin II-B during mouse brain development. Mol Biol Cell. 2006;17:2138–49.
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–7.
Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, Weiss ME, Köster J, Marais A, et al. Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet. 2017;25:176–82.
Scheidecker S, Etard C, Pierce NW, Geoffroy V, Schaefer E, Muller J, et al. Exome sequencing of Bardet-Biedl syndrome patient identifies a null mutation in the BBSome subunit BBIP1 (BBS18). J Med Genet. 2014;51:132–6.
Müller F, Blader P, Rastegar S, Fischer N, Knöchel W, Strähle U. Characterization of zebrafish smad1, smad2 and smad5: the amino-terminus of smad1 and smad5 is required for specific function in the embryo. Mech Dev. 1999;88:73–88. Oct
Yang L, Rastegar S, Strähle U. Regulatory interactions specifying Kolmer-Agduhr interneurons. Development. 2010;137:2713–22.
Armant O, März M, Schmidt R, Ferg M, Diotel N, Ertzer R, et al. Genome-wide, whole mount in situ analysis of transcriptional regulators in zebrafish embryos. Dev Biol. 2013;380:351–62.
Furniss D, Kan SH, Taylor IB, Johnson D, Critchley PS, Giele HP, et al. Genetic screening of 202 individuals with congenital limb malformations and requiring reconstructive surgery. J Med Genet. 2009;46:730–5.
Wilson CA, Tsuchida MA, Allen GM, Barnhart EL, Applegate KT, Yam PT, et al. Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature. 2010;465:373–7.
Huang Y, Wang X, Wang X, Xu M, Liu M, Liu D. Nonmuscle myosin II-B (myh10) expression analysis during zebrafish embryonic development. Gene Expr Patterns. 2013;13:265–70.
Gutzman JH, Sahu SU, Kwas C. Non-muscle myosin IIA and IIB differentially regulate cell shape changes during zebrafish brain morphogenesis. Dev Biol. 2015;397:103–15.
Lusk S, Kwan KM. Pax2a, but not pax2b, influences cell survival and periocular mesenchyme localization to facilitate zebrafish optic fissure closure. Dev Dyn. 2022;251:625–44.
Rivière JB, van Bon BWM, Hoischen A, Kholmanskikh SS, O’Roak BJ, Gilissen C, et al. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat Genet. 2012;44:440–4.
Haviv L, Gillo D, Backouche F, Bernheim-Groswasser A. A cytoskeletal demolition worker: myosin II acts as an actin depolymerization agent. J Mol Biol. 2008;375:325–30.
Berger J, Sztal T, Currie PD. Quantification of birefringence readily measures the level of muscle damage in zebrafish. Biochem Biophys Res Commun. 2012;423:785–8.
Hall TE, Bryson-Richardson RJ, Berger S, Jacoby AS, Cole NJ, Hollway GE, et al. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc Natl Acad Sci USA. 2007;104:7092–7.
Charvet B, Guiraud A, Malbouyres M, Zwolanek D, Guillon E, Bretaud S, et al. Knockdown of col22a1 gene in zebrafish induces a muscular dystrophy by disruption of the myotendinous junction. Development. 2013;140:4602–13.
Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4). Am J Hum Genet. 2004;74:770–6.
Heath KE, Campos-Barros A, Toren A, Rozenfeld-Granot G, Carlsson LE, Savige J, et al. Nonmuscle myosin heavy chain IIA mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am J Hum Genet. 2001;69:1033–45.
Garrido-Casado M, Asensio-Juárez G, Vicente-Manzanares M. Nonmuscle Myosin II regulation directs its multiple roles in cell migration and division. Annu Rev Cell Dev Biol. 2021;37:285–310.
Ivanov AI, Lechuga S, Marino-Melendez A, Naydenov NG. Unique and redundant functions of cytoplasmic actins and nonmuscle myosin II isoforms at epithelial junctions. Ann N. Y Acad Sci. 2022;1515:61–74.
Diacou R, Nandigrami P, Fiser A, Liu W, Ashery-Padan R, Cvekl A. Cell fate decisions, transcription factors and signaling during early retinal development. Prog Retin Eye Res. 2022;91:101093.
Wei Q, Adelstein RS. Conditional expression of a truncated fragment of nonmuscle myosin II-A alters cell shape but not cytokinesis in HeLa cells. Mol Biol Cell. 2000;11:3617–27.
Ma X, Adelstein RS. The role of vertebrate nonmuscle Myosin II in development and human disease. Bioarchitecture. 2014;4:88–102.
Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65.
Veigel C, Schmidt CF. Moving into the cell: single-molecule studies of molecular motors in complex environments. Nat Rev Mol Cell Biol. 2011;12:163–76.
Tuzovic L, Yu L, Zeng W, Li X, Lu H, Lu HM, et al. A human de novo mutation in MYH10 phenocopies the loss of function mutation in mice. Rare Dis. 2013;1:e26144.
Hamdan FF, Srour M, Capo-Chichi JM, Daoud H, Nassif C, Patry L, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772.
Jurgens JA, Barry BJ, Chan WM, MacKinnon S, Whitman MC, Matos Ruiz PM, et al. Expanding the genetics and phenotypes of ocular congenital cranial dysinnervation disorders. Genet Med. 2024;101216. https://doi.org/10.1016/j.gim.2024.101216.
Yates TM, Turner CL, Firth HV, Berg J, Pilz DT. Baraitser-Winter cerebrofrontofacial syndrome. Clin Genet. 2017;92:3–9.
Verloes A, Di Donato N, Masliah-Planchon J, Jongmans M, Abdul-Raman OA, Albrecht B, et al. Baraitser-Winter cerebrofrontofacial syndrome: delineation of the spectrum in 42 cases. Eur J Hum Genet. 2015;23:292–301.
Yang Q, Zhang XF, Pollard TD, Forscher P. Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J Cell Biol. 2012;197:939–56.
MacArthur MW, Thornton JM. Influence of proline residues on protein conformation. J Mol Biol. 1991;218:397–412.
Schimmel PR, Flory PJ. Conformational energies and configurational statistics of copolypeptides containing L-proline. J Mol Biol. 1968;34:105–20.
Richardson R, Tracey-White D, Webster A, Moosajee M. The zebrafish eye-a paradigm for investigating human ocular genetics. Eye. 2017;31:68–86.
Kim HT, Yin W, Jin YJ, Panza P, Gunawan F, Grohmann B, et al. Myh10 deficiency leads to defective extracellular matrix remodeling and pulmonary disease. Nat Commun. 2018;9:4600.
Myhre JL, Hills JA, Jean F, Pilgrim DB. Unc45b is essential for early myofibrillogenesis and costamere formation in zebrafish. Dev Biol. 2014;390:26–40.
Sparrow JC, Schöck F. The initial steps of myofibril assembly: integrins pave the way. Nat Rev Mol Cell Biol. 2009;10:293–8.
Sharp WW, Simpson DG, Borg TK, Samarel AM, Terracio L. Mechanical forces regulate focal adhesion and costamere assembly in cardiac myocytes. Am J Physiol. 1997;273:H546–556.
Kröll-Hermi A. Identification and validation of novel genes implicated in neurosensory and neurological diseases [Internet]. (2019). Available from: https://theses.fr/2019STRAJ119.
Verloes A, Drunat S, Pilz D, Di Donato N. Baraitser-Winter Cerebrofrontofacial Syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJ, Stephens K, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; (1993) [cited 2019 Nov 25]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK327153/.
Acknowledgements
We wish to warmly thank the families for their participation. We would like to thank Alain Verloes (Hôpital Robert-Debré, Paris, France), Nicola Ragge (Oxford Brookes University, Oxford, UK) and Patrick Calvas (CHU Toulouse, France) as well as the ERN-Eye network for attempting to identify patients with the same phenotype. The authors would like to thank the BiGEst-ICube Platform for assistance. This research formed part of a doctoral dissertation by Ariane Kröll-Hermi, entitled “Identification and Validation of novel genes implicated in Neurosensory and Neurological Diseases” [44, 45].
Funding
We would like to thank Jean-Louis Mandel, the financial support of the Centre Régional de Génétique Médicale de Strasbourg and the Caisse d’Assurance Retraite et de la Santé au Travail Alsace-Moselle. AK-H was supported by a doctoral fellowship from the Initiatives d’Excellence (IdEx) through the University of Strasbourg and by the Franco-German University (UFA/DFH). SB and SF were supported by INSERM and grants from the Initiatives d’Excellence (IdEx, Université de Strasbourg) and the AFM-Téléthon. Sequencing was performed by the GenomEast platform, a member of the “France Génomique” consortium (ANR-10-INBS-0009).
Author information
Authors and Affiliations
Contributions
HD and SF provided direction for the project, conceived and designed the experiments; BR performed cloning and mutagenesis experiments; SB, CS, CD, SS, SaS, and CJ performed cell biology experiments and data analyses; SS, ES, XZ, AK, ER, and HD, gathered data from patients and performed clinical investigations; AK-H, CE, OK, and US designed and performed the zebrafish experiments and data analyses; VG, J-BL, and JM gathered sequencing data and performed analyses for families 1 and 3; AB-A and EZ. analysed the genetic data for Family 2; HD, SF, SS, and SB analysed the data and wrote the paper. All authors critically revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This research followed the tenets of the Declaration of Helsinki. Approval was obtained from the institutional review boards “Comité de Protection des Personnes” (EST IV, N◦DC-20142222), Northern Ostrobothnia Hospital District, Oulu, Finland (EETTMK: 45/2015, amendment 2020), and Strasbourg University Hospital ethics committee as part of the ultra-rare disease cohort. Informed consent was obtained before study inclusion. Written informed consent for publication of images was also obtained.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Scheidecker, S., Bär, S., Kröll-Hermi, A. et al. Novel MYH10 heterozygous variants associated to a syndrome combining mainly ptosis and ocular coloboma expand the MYH10 related phenotypes. Eur J Hum Genet 33, 1432–1441 (2025). https://doi.org/10.1038/s41431-025-01803-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-025-01803-2
This article is cited by
-
Mutations in CFAP57 disrupt the localization of MYH10 and IFT88, leading to flagellogenesis failure in humans and mice
Human Genomics (2025)
-
Insights in genetics: from molecular mechanisms to patient perspectives
European Journal of Human Genetics (2025)