Abstract
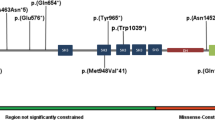
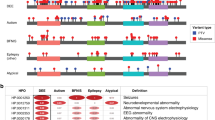
The CCR4-NOT complex, crucial in gene expression regulation, includes CNOT3, a subunit linked to neurodevelopmental disorders when mutated. This study investigates 51 patients from 42 families with heterozygous CNOT3 variants, aiming to expand the understanding of CNOT3-related neurodevelopmental disorders and explore genotype-phenotype correlations. Patients originated from various countries, reflecting the disorder’s global significance. All patients exhibited developmental delays, particularly in the language area. Intellectual disability was found in 87% of patients and was typically mild to moderate. Behavioral issues, including autism spectrum disorders and attention deficits, were common, affecting over half of the patients. Dysmorphic features were highlighted and may help establishing the diagnosis. Epilepsy was uncommon (10%). Twenty-eight novel variants were identified, including missense, nonsense, frameshift, intronic variations and a deletion of 12 exons. Missense variants clustered at the N- and C-terminal regions of the protein, indicating critical functional roles. No clear genotype-phenotype correlation was observed, suggesting that all identified variants resulted in a loss-of-function effect. Finally, this work delineates the clinical and molecular spectrum of CNOT3-related disorders thanks to an in-depth characterization of a large cohort. Further research will be necessary to understand the functional consequences of the variants and enhance patient long-term outcomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request. Pathogenic and probably pathogenic variants in our cohort that had not previously been reported were submitted to ClinVar (Supplementary Data S3).
References
Collart MA. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip Rev RNA. 2016;7:438–54.
Martin R, Splitt M, Genevieve D, Aten E, Collins A, de Bie CI, et al. De novo variants in CNOT3 cause a variable neurodevelopmental disorder. Eur J Hum Genet. 2019;27:1677–82.
Meyer R, Begemann M, Demuth S, Kraft F, Dey D, Schüler H, et al. Inherited cases of CNOT3-associated intellectual developmental disorder with speech delay, autism, and dysmorphic facies. Clin Genet. 2020;98:408–12.
Pinard A, Guey S, Guo D, Cecchi AC, Kharas N, Wallace S, et al. The pleiotropy associated with de novo variants in CHD4, CNOT3, and SETD5 extends to moyamoya angiopathy. Genet Med J Am Coll Med Genet. 2020;22:427–31.
Priolo M, Radio FC, Pizzi S, Pintomalli L, Pantaleoni F, Mancini C, et al. Co-Occurring Heterozygous CNOT3 and SMAD6 Truncating Variants: Unusual Presentation and Refinement of the IDDSADF Phenotype. Genes. 2021;12:1009
Lv J, Ming WJ, Zheng Y, Xu S, Fang GL, Zhang Q, et al. A novel pathogenic variant in CNOT3 causing neurodevelopmental delay and epilepsy. Seizure. 2023;107:104–6.
Zhao P, Meng Q, Wan C, Lei T, Zhang L, Zhang X, et al. Clinical features of CNOT3-associated neurodevelopmental disorder in three Chinese patients. Neurogenet. 2023;24:129–36.
Lee CG, Kim HJ, Seol CA, Ki CS. Novel in-frame deletion CNOT3 variant in a family with intellectual developmental disorder with speech delay and dysmorphic facies. Neurol Genet. 2024;10:e200116.
Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30.
DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation - PMC [Internet]. [cité 1 sept 2023]. Disponible sur: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3965078/.
Baux D, Van Goethem C, Ardouin O, Guignard T, Bergougnoux A, Koenig M, et al. MobiDetails: online DNA variants interpretation. Eur J Hum Genet EJHG. 2021;29:356–60.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–94.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–7.
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–7.
Fokkema IFAC, den Dunnen JT, Taschner PEM. LOVD: easy creation of a locus-specific sequence variation database using an « LSDB-in-a-box » approach. Hum Mutat. 2005;26:63–8.
Day INM. dbSNP in the detail and copy number complexities. Hum Mutat. 2010;31:2–4.
Gudmundsson S, Singer-Berk M, Watts NA, Phu W, Goodrich JK, Solomonson M, et al. Variant interpretation using population databases: Lessons from gnomAD. Hum Mutat. 2022;43:1012–30.
Nassar LR, Barber GP, Benet-Pagès A, Casper J, Clawson H, Diekhans M, et al. The UCSC Genome Browser database: 2023 update. Nucleic Acids Res. 2022;51:D1188–95.
Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010;6:e1001025.
Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4.
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–7.
Leman R, Parfait B, Vidaud D, Girodon E, Pacot L, Le Gac G, et al. SPiP: Splicing Prediction Pipeline, a machine learning tool for massive detection of exonic and intronic variant effects on mRNA splicing. Hum Mutat. 2022;43:2308–23.
罕见病数据库 [Internet]. [cité 2 févr 2025]. Disponible sur: https://rddc.tsinghua-gd.org/tool/rna-splicer?id=4849.
de Sainte Agathe JM, Filser M, Isidor B, Besnard T, Gueguen P, Perrin A, et al. SpliceAI-visual: a free online tool to improve SpliceAI splicing variant interpretation. Hum Genomics. 2023;17:7.
Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015;31:3359–61.
on behalf of the ACMG Laboratory Quality Assurance Committee, Richards S, Aziz N, Bale S, Bick D, Das S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.
Franklin by Genoox: The Future of Genomic Medicine. 2022. [Internet]. 2022. Disponible sur: https://franklin.genoox.com.
Rodrigues CHM, Pires DEV, Ascher DB. DynaMut2: assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci Publ Protein Soc. 2021;30:60–9.
Boland A, Chen Y, Raisch T, Jonas S, Kuzuoğlu-Öztürk D, Wohlbold L, et al. Structure and assembly of the NOT module of the human CCR4-NOT complex. Nat Struct Mol Biol. 2013;20:1289–97.
Collart MA, Panasenko OO, Nikolaev SI. The Not3/5 subunit of the Ccr4-Not complex: A central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell Signal. 2013;25:743–51.
Absmeier E, Chandrasekaran V, O’Reilly FJ, Stowell JA, Rappsilber J, Passmore LA. Specific recognition and ubiquitination of translating ribosomes by mammalian CCR4-NOT. Nat Struct Mol Biol. 2023;30:1314.
Wiel L, Baakman C, Gilissen D, Veltman JA, Vriend G, Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum Mutat. 2019;40:1030–8.
Funding
Research reported in this publication was supported by Jordan’s Guardian Angels, the Sunderland Foundation and the Brotman Baty Institute (to G.M.M.). E.W. : The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003]. This study makes use of DECIPHER (http://www.deciphergenomics.org), which is funded by Wellcome [grant number WT223718/Z/21/Z]. See Nature PMID: 25533962 or www.ddduk.org/access.html for full acknowledgement. P.Z.’s work is being supported by Stiftung Michael through the generous assistance of the Canger-Janz-Fellowship. L.N. was supported by the project National Institute for Neurological Research (Programme EXCELES, ID Project No. LX22NPO5107) - Funded by the E’uropean Union – Next Generation EU. L.N. and S.K. were supported by grant NU23-07-00281 from the Ministry of Health of the Czech Republic, by the institutional program UNCE/24/MED/022 of Charles University in Prague and they thank to the National Center for Medical Genomics (LM2023067) for WES analyses. This work was partially funded by the Italian Ministry of Health RC2025/2026.
Author information
Authors and Affiliations
Contributions
CE, MR, JA collected clinical and molecular data. CE, JP analyzed and interpreted data and wrote the manuscript. JP designed the study. EA, FA, ALAD, EKB, PB, LPB, BC, VC, MC, BC, SC, CC, BD, ED, ADD, ASDP, BD, LD, JF, DG, TG, MMH, DH, IH, MI, BI, BK, SK, DAK, AK, JL, CL, JL, CL, SM, SM, CM, GM, IN, SN, LN, EP, AP, JP, JP, FP, AR, AR, MR, AR, LR, MS, JS, EHS, AS, RS, TS, JS, PS, MS, HS, FTMT, AMT, JVG, GV, JJAV, CV, CVD, EV, EW, PZ, FZ, PK, JP characterized the clinical and molecular features of the disease. All the authors edited or commented the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Individuals were referred by clinical geneticists for genetic testing as part of routine clinical care. All patients enrolled and/or their legal representative have signed informed consent for use of data and/or photographs.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Engel, C., Rendek, M., Assoumani, J. et al. Comprehensive analysis of CNOT3-related neurodevelopmental disorders: phenotypic and genotypic characterization. Eur J Hum Genet 33, 989–996 (2025). https://doi.org/10.1038/s41431-025-01884-z
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-025-01884-z
This article is cited by
-
Genomic medicine in full bloom: a summer farewell issue
European Journal of Human Genetics (2025)