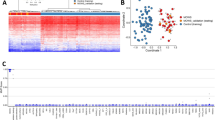
Abstract
Neurodevelopmental disorder with hypotonia, stereotypic hand movements, and impaired language (NEDHSIL), also known as MEF2C-related disorder or MEF2C haploinsufficiency syndrome (MCHS), is a condition caused by pathogenic variants in the Myocyte Enhancer Factor-2C (MEF2C) gene. This study aimed to identify a DNA methylation episignature specific to NEDHSIL and explore its similarities with other known episignatures. Genome-wide DNA levels were assessed in a cohort of patients with MEF2C mutations and controls, and differentially methylated CpG sites were identified. A bioinformatic analysis yielded a classifier that was trained against controls and other known episignature disorders within the EpiSign Knowledge Database. The classifier demonstrated high accuracy, sensitivity, and specificity in classifying NEDHSIL samples. Furthermore, functional annotation and comparative analysis revealed similarities between the NEDHSIL episignature and other genetic neurodevelopmental disorders. This study provides evidence for a DNA methylation episignature specific to NEDHSIL and highlights the potential utility of this epigenetic biomarker for diagnosing and understanding molecular pathophysiology of neurodevelopmental disorders associated with MEF2C mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Datasets used in this study that are available publicly are previously described [23]. Anonymized data for each subject is described in the study. The individual genomic and epigenomic or any other personally identifiable data for other samples in the EpiSign Knowledge Database (EKD) are not available for deposition in publicly accessible databases due to institutional and ethics restrictions. Specifically, these include data and samples submitted from external institutions to London Health Sciences EKD that are subject to Institutional Material and Data Transfer agreements, data submitted to London Health Sciences for episignature assessment under Research Services Agreements, and research study cohorts under Institutional Research Ethics Approval (Western University REB 106302; and REB 116108). Some of the software packages used in this study are publicly available as described in the Materials and Methods. EpiSignTM is a commercial software and is not publicly available.
References
Vrečar I, Innes J, Jones E, Kingston H, Reardon W, Kerr B, et al. Further Clinical Delineation of the MEF2C Haploinsufficiency Syndrome: Report on New Cases and Literature Review of Severe Neurodevelopmental Disorders Presenting with Seizures, Absent Speech, and Involuntary Movements. J Pediatr Genet. 2017;06:129–41. https://doi.org/10.1055/s-0037-1601335.
Paciorkowski AR, Traylor RN, Rosenfeld JA, Hoover JM, Harris CJ, Winter S, et al. MEF2C Haploinsufficiency features consistent hyperkinesis, variable epilepsy, and has a role in dorsal and ventral neuronal developmental pathways. Neurogenetics. 2013;14:99–111. https://doi.org/10.1007/s10048-013-0356-y.
Adrião A, Santana I, Ribeiro C, Cancela ML, Conceição N, Grazina M. Identification of a novel mutation in MEF2C gene in an atypical patient with frontotemporal lobar degeneration. Neurological Sci. 2022;43:319–26. https://doi.org/10.1007/s10072-021-05269-0.
Bienvenu T, Diebold B, Chelly J, Isidor B. Refining the phenotype associated with MEF2C point mutations. Neurogenetics. 2013;14:71–5. https://doi.org/10.1007/s10048-012-0344-7.
Wen F, Hou J, Ji X, Chu X, Liu X, Shi Z, et al. The Mef2c/AdipoR1 axis is responsible for myogenic differentiation and is regulated by resistin in skeletal muscles. Gene. 2023;857:147193. https://doi.org/10.1016/j.gene.2023.147193.
Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, et al. Integrated Transcript and Genome Analyses Reveal NKX2-1 and MEF2C as Potential Oncogenes in T Cell Acute Lymphoblastic Leukemia. Cancer Cell. 2011;19:484–97. https://doi.org/10.1016/j.ccr.2011.02.008.
Orozco Morales ML, Rinaldi CA, de Jong E, Lansley SM, Lee YCG, Zemek RM, et al. Geldanamycin treatment does not result in anti-cancer activity in a preclinical model of orthotopic mesothelioma. PLoS ONE. 2023;18:e0274364. https://doi.org/10.1371/journal.pone.0274364.
Haberland M, Arnold MA, McAnally J, Phan D, Kim Y, Olson EN. Regulation of HDAC9 Gene Expression by MEF2 Establishes a Negative-Feedback Loop in the Transcriptional Circuitry of Muscle Differentiation. Mol Cell Biol. 2007;27:518–25. https://doi.org/10.1128/MCB.01415-06.
Cosgrove D, Whitton L, Fahey L, Ó Broin P, Donohoe G, Morris DW. Genes influenced by MEF2C contribute to neurodevelopmental disease via gene expression changes that affect multiple types of cortical excitatory neurons. Hum Mol Genet. 2021;30:961–70. https://doi.org/10.1093/hmg/ddaa213.
Zweier M, Rauch A. The MEF2C-Related and 5q14.3q15 Microdeletion Syndrome. Mol Syndromol. 2011;2:164–70. https://doi.org/10.1159/000337496.
Lambert L, Bienvenu T, Allou L, Valduga M, Echenne B, Diebold B, et al. MEF2C mutations are a rare cause of Rett or severe Rett-like encephalopathies. Clin Genet. 2012;82:499–501. https://doi.org/10.1111/j.1399-0004.2012.01861.x.
Rocha H, Sampaio M, Rocha R, Fernandes S, Leão M. MEF2C haploinsufficiency syndrome: Report of a new MEF2C mutation and review. Eur J Med Genet. 2016;59:478–82. https://doi.org/10.1016/j.ejmg.2016.05.017.
Bjornsson HT. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473–81. https://doi.org/10.1101/gr.190629.115.
der Laan L, van, Rooney K, Trooster TM, Mannens MM, Sadikovic B, Henneman P. DNA methylation episignatures: insight into copy number variation. Epigenomics. 2022;14:1373–88. https://doi.org/10.2217/epi-2022-0287.
Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–40. https://doi.org/10.1242/dev.008367.
Zhang Z, Zhao Y. Progress on the roles of MEF2C in neuropsychiatric diseases. Mol Brain. 2022;15:8. https://doi.org/10.1186/s13041-021-00892-6.
Flavell SW, Kim T-K, Gray JM, Harmin DA, Hemberg M, Hong EJ, et al. Genome-Wide Analysis of MEF2 Transcriptional Program Reveals Synaptic Target Genes and Neuronal Activity-Dependent Polyadenylation Site Selection. Neuron. 2008;60:1022–38. https://doi.org/10.1016/j.neuron.2008.11.029.
Levy MA, McConkey H, Kerkhof J, Barat-Houari M, Bargiacchi S, Biamino E, et al. Novel diagnostic DNA methylation episignatures expand and refine the epigenetic landscapes of Mendelian disorders. Human Genet Genomics Adv. 2022;3:100075. https://doi.org/10.1016/j.xhgg.2021.100075.
Aref-Eshghi E, Rodenhiser DI, Schenkel LC, Lin H, Skinner C, Ainsworth P, et al. Genomic DNA Methylation Signatures Enable Concurrent Diagnosis and Clinical Genetic Variant Classification in Neurodevelopmental Syndromes. Am J Hum Genet. 2018;102:156–74. https://doi.org/10.1016/j.ajhg.2017.12.008.
Kerkhof J, Rastin C, Levy MA, Relator R, McConkey H, Demain L, et al. Diagnostic utility and reporting recommendations for clinical DNA methylation episignature testing in genetically undiagnosed rare diseases. Genet Med 2024:101075. https://doi.org/10.1016/j.gim.2024.101075.
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–9. https://doi.org/10.1093/bioinformatics/btu049.
Aref-Eshghi E, Bend EG, Colaiacovo S, Caudle M, Chakrabarti R, Napier M, et al. Diagnostic Utility of Genome-wide DNA Methylation Testing in Genetically Unsolved Individuals with Suspected Hereditary Conditions. Am J Hum Genet. 2019;104:685–700. https://doi.org/10.1016/j.ajhg.2019.03.008.
Aref-Eshghi E, Kerkhof J, Pedro VP, Barat-Houari M, Ruiz-Pallares N, Andrau J-C, et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. American J Hum Genet. 2020;106:356–70. https://doi.org/10.1016/j.ajhg.2020.01.019.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47. https://doi.org/10.1093/nar/gkv007.
Ho DE, Imai K, King G, Stuart EA. MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. J Stat Softw 2011;42. https://doi.org/10.18637/jss.v042.i08.
Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinforma. 2012;13:86. https://doi.org/10.1186/1471-2105-13-86.
Levy MA, Relator R, McConkey H, Pranckeviciene E, Kerkhof J, Barat-Houari M, et al. Functional correlation of genome-wide DNA methylation profiles in genetic neurodevelopmental disorders. Hum Mutat. 2022;43:1609–28. https://doi.org/10.1002/humu.24446.
Kolde, R. pheatmap: Pretty Heatmaps. R package version 1.0.12; 2019. Available from: https://CRAN.R-project.org/package=pheatmap.
Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2. https://doi.org/10.1093/bioinformatics/btu393.
Cavalcante RG, Sartor MA. annotatr: genomic regions in context. Bioinformatics. 2017;33:2381–3. https://doi.org/10.1093/bioinformatics/btx183.
Cardoso MA, Rizzardi LEA, Kume LW, Groeneveld CS, Trefflich S, Morais DAA, et al. TreeAndLeaf: an R/Bioconductor package for graphs and trees with focus on the leaves. Bioinformatics. 2022;38:1463–4. https://doi.org/10.1093/bioinformatics/btab819.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics Med. 2015;17:405–24. https://doi.org/10.1038/gim.2015.30.
Haghshenas S, Foroutan A, Bhai P, Levy MA, Relator R, Kerkhof J, et al. Identification of a DNA methylation signature for Renpenning syndrome (RENS1), a spliceopathy. Eur J Human Genet 2023. https://doi.org/10.1038/s41431-023-01313-z.
Vos N, Reilly J, Elting MW, Campeau PM, Coman D, Stark Z, et al. DNA methylation episignatures are sensitive and specific biomarkers for detection of patients with KAT6A / KAT6B variants. Epigenomics 2023. https://doi.org/10.2217/epi-2023-0079.
Rooney K, van der Laan L, Trajkova S, Haghshenas S, Relator R, Lauffer P, et al. DNA methylation episignature and comparative epigenomic profiling of HNRNPU-related neurodevelopmental disorder. Genetics Med. 2023;25:100871. https://doi.org/10.1016/j.gim.2023.100871.
Kojima H, Sadahiro T, Muraoka N, Yamakawa H, Hashimoto H, Ishii R, et al. MEF2C/p300-mediated epigenetic remodeling promotes the maturation of induced cardiomyocytes. Stem Cell Rep. 2023;18:1274–83. https://doi.org/10.1016/j.stemcr.2023.05.001.
Aref-Eshghi E, Schenkel LC, Lin H, Skinner C, Ainsworth P, Paré G, et al. The defining DNA methylation signature of Kabuki syndrome enables functional assessment of genetic variants of unknown clinical significance. Epigenetics. 2017;12:923–33. https://doi.org/10.1080/15592294.2017.1381807.
van Jaarsveld RH, Reilly J, Cornips M-C, Hadders MA, Agolini E, Ahimaz P, et al. Delineation of a KDM2B-related neurodevelopmental disorder and its associated DNA methylation signature. Genetics Med. 2023;25:49–62. https://doi.org/10.1016/j.gim.2022.09.006.
Levy MA, Beck DB, Metcalfe K, Douzgou S, Sithambaram S, Cottrell T, et al. Deficiency of TET3 leads to a genome-wide DNA hypermethylation episignature in human whole blood. NPJ Genom Med. 2021;6:92 https://doi.org/10.1038/s41525-021-00256-y.
Uchino S, Waga C. SHANK3 as an autism spectrum disorder-associated gene. Brain Dev. 2013;35:106–10. https://doi.org/10.1016/j.braindev.2012.05.013.
Wright CF, Quaife NM, Ramos-Hernández L, Danecek P, Ferla MP, Samocha KE, et al. Non-coding region variants upstream of MEF2C cause severe developmental disorder through three distinct loss-of-function mechanisms. Am J Hum Genet. 2021;108:1083–94. https://doi.org/10.1016/j.ajhg.2021.04.025.
Basu S, Ro EJ, Liu Z, Kim H, Bennett A, Kang S, et al. The Mef2c Gene Dose-Dependently Controls Hippocampal Neurogenesis and the Expression of Autism-Like Behaviors. J Neurosci 2024;44. https://doi.org/10.1523/JNEUROSCI.1058-23.2023.
Bai G, Yuan R, Yuan J, Liu Y, Zhao S, Zhang X. A rare Coffin-Siris syndrome induced by SOX11: a de novo nonsense variant of short stature. BMC Med Genomics. 2024;17:1–9. https://doi.org/10.1186/S12920-024-02036-W/TABLES/2.
Ding Y, Chen J, Tang Y, Chen LN, Yao RE, Yu T, et al. Identification and functional analysis of novel SOX11 variants in Chinese patients with Coffin-Siris syndrome 9. Front Genet 2022;13. https://doi.org/10.3389/FGENE.2022.940776.
Li J, Yu Y, Zhao J, Zhang J, Wang Y, Ding K, et al. Genetic variants of the type-3 metabotropic glutamate receptor gene associated with human spatial localization ability. Gene Rep. 2021;23:101135. https://doi.org/10.1016/j.genrep.2021.101135.
Bonnet M, Roche F, Fagotto-Kaufmann C, Gazdagh G, Truong I, Comunale F, et al. Pathogenic TRIO variants associated with neurodevelopmental disorders perturb the molecular regulation of TRIO and axon pathfinding in vivo. Mol Psychiatry. 2023;28:1527–44. https://doi.org/10.1038/s41380-023-01963-x.
Zhu S, Tao M, Li Y, Wang X, Zhao Z, Liu Y, et al. H3K27me3 of Rnf19a promotes neuroinflammatory response during Japanese encephalitis virus infection. J Neuroinflammat. 2023;20:168. https://doi.org/10.1186/s12974-023-02852-4.
Acknowledgements
We extend our gratitude to the participants and their families for their invaluable contribution.
Funding
Funding for this study is provided in part by the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-188).
Author information
Authors and Affiliations
Contributions
Conceptualization: AS, BS; Data curation: AS JK; Formal analysis: AS, SH, MAL, RR; Funding acquisition: BS; Sample Collection: SAS, AV, IV, IES, KAM, MLT, JACC; Investigation: AS, MAL, JK, RR; Methodology: AS, MAL, RR; Project administration: HM, BS; Software: AS, MAL, RR; Supervision: BS; Validation: AS, SH, LVDL; Visualization: AS; Writing-original draft: AS; Writing-review and editing: AS, LVDL, SH, BS, JACC.
Corresponding author
Ethics declarations
Competing interests
BS is a shareholder in EpiSign Inc involved in commercial applications of EpiSign™ technology.
Ethical approval
All samples and data records were anonymized to protect the identities of the individuals involved. The research protocol received ethical approval from the Research Ethics Board (REB 106302) at Western University and the Institutional Review Board (IRB) of Self Regional Healthcare. Informed consent was obtained from physicians for the utilization of clinical information pertaining to the described patients.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Silva, A., Haghshenas, S., van der Laan, L. et al. Identification of an episignature for the MEF2C-associated syndrome. Eur J Hum Genet 34, 53–60 (2026). https://doi.org/10.1038/s41431-025-01983-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-025-01983-x
This article is cited by
-
New year, new insights in genomic medicine
European Journal of Human Genetics (2026)