Abstract
Elucidating the factors that drive the genetic patterns of natural populations is key in evolutionary biology, ecology and conservation. Hence, it is crucial to understand the role that environmental features play in species genetic diversity and structure. Landscape genetics measures functional connectivity and evaluates the effects of landscape composition, configuration, and heterogeneity on microevolutionary processes. Deserts constitute one of the world’s most widespread biomes and exhibit a striking heterogeneity of microhabitats, yet few landscape genetics studies have been performed with rodents in deserts. We evaluated the relationship between landscape and functional connectivity, at a microgeographic scale, of the Nelson’s pocket mouse Chaetodipus nelsoni in the Mapimí Biosphere Reserve (Chihuahuan desert). We used single-nucleotide polymorphisms and characterized the landscape based on on-site environmental data and from Landsat satellite images. We identified two distinct genetic clusters shaped by elevation, vegetation and soil. High elevation group showed higher connectivity in the elevated zones (1250–1350 m), with scarce vegetation and predominantly rocky soils; whereas that of Low elevation group was at <1200 m, with denser vegetation and sandy soils. These genetic patterns are likely associated with the species’ locomotion type, feeding strategy and building of burrows. Interestingly, we also identified morphological differences, where hind foot size was significantly smaller in individuals from High elevation compared to Low elevation, suggesting the possibility of ecomorphs associated with habitat differences and potential local adaptation processes, which should be explored further. These findings improve our understanding of the genetics and ecology of C. nelsoni and other desert rodents.
Similar content being viewed by others
Introduction
Biological diversity in its finest level is defined by genetic diversity, the intra-specific variability essential for the ability of individuals to survive environmental and habitat changes, the viability of populations and the evolution and adaptation of species (Primack 2014; Mimura et al. 2017; Hoban et al. 2020). In turn, life history traits, environmental conditions and landscape characteristics influence genetic diversity and structure. Functional connectivity, the extent to which a landscape facilitates or impedes the dispersal (gene flow) of individuals across the landscape, greatly determines the genetic diversity and structure of populations (Spear et al. 2015; Peterman 2018). Both natural (e.g. environmental variables, habitat features, resources availability) and anthropogenic factors (e.g. land use changes, habitat modification, fragmentation) influence the behaviour and movement of individuals (Fahrig 2003; Manel et al. 2003; Balkenhol et al. 2016), also shaping genetic patterns (Cushman et al. 2006; Frankham 2010; LaPoint et al. 2015). Thereby, disentangling the landscape factors that drive functional connectivity of natural populations is key in evolutionary biology, ecology and conservation.
Landscape genetics measures functional connectivity to uncover the effects of landscape composition, configuration, and heterogeneity on microevolutionary processes (Manel et al. 2003; Schoville et al. 2012; Balkenhol et al. 2016). Landscape variables are typically modelled as isolation by environment and measured as resistance (Wang and Bradburd 2014; Balkenhol et al. 2016; Flores-Manzanero et al. 2019). Resistance surfaces are spatial layers with values that denote the degree to which a landscape feature restricts or facilitates connectivity; thus, resistance surfaces can be viewed as hypotheses of the relationship between landscape variables and gene flow (Spear et al. 2015; Peterman et al. 2014; Peterman 2018). Landscape genetics studies performed with small rodents have identified variables like precipitation, vegetation cover, elevation and temperature to be significant drivers of functional connectivity in natural and modified landscapes (Castillo et al. 2014; Marrotte et al. 2014; Mullins et al. 2015; Garrido-Garduño et al. 2016; Russo et al. 2016; Flores-Manzanero et al. 2019; Borja-Martínez et al. 2022).
Deserts constitute one of the most widespread biomes in the world, occupying approximately one third of the Earth’s surface. These ecosystems exhibit a striking heterogeneity of microhabitats where vegetation tends to be distributed in small patches. They encompass diverse spatial and temporal scales and harbour a great diversity of taxonomic groups, many of which are endemic (Whitford 2002; WWF World Wildlife Fund (2019)). Hence, these ecosystems are ideal to evaluate hypotheses about animal dispersal and functional connectivity under the context of landscape genetics (Manel and Holderegger 2013; Garrido-Garduño et al. 2022). Nonetheless, very few landscape genetics studies have been performed with rodents in deserts of the Americas in general (Storfer et al. 2010; Flores-Manzanero and Vázquez-Domínguez 2019) and in Mexican deserts in particular (Cosentino et al. 2015; Flores-Manzanero et al. 2019). Moreover, much less research has been performed considering the genetic structuring of desert rodent populations at microgeographic scales, i.e. landscape scale pertaining to their small size, short-distance dispersal and harsh environmental conditions of their habitat (low productivity, extreme temperatures and low precipitation regimes) (Flores-Manzanero et al. 2019; Flores-Manzanero and Vázquez-Domínguez 2019).
Chaetodipus is a genus of pocket mice that includes 17 species endemic to arid environments in North America and Mexico (Vaughan et al. 2000). The Nelson’s pocket mouse, Chaetodipus nelsoni, has a wide distribution along the Chihuahuan desert, from central-north Mexico to north-western Texas and southeast New Mexico (Best 1994; Patton 2005), including the Mapimí Biosphere Reserve (MBR) in Mexico. It is a medium-sized rodent (total length 180 mm), with a long tail (102 mm), long hind feet (ca. 30% of the length of head and body), and with characteristic cheek pouches and tufted-tip tails. Little is known about C. nelsoni population genetics; most published studies describe its taxonomy and systematics, distribution and morphology from expeditions done by last mid-century naturalists (Best 1994; Patton 1970, 2005; Patton et al. 1981; Modi 2003), and more recently about some ecological aspects (Geluso and Geluso 2015) and phylogeography (Neiswenter et al. 2019). From these works we can gather that it does not have sexual dimorphism, primarily feeds on seeds, is active all year, and builds burrows at the bases of certain plant species using mainly the hind feet. The type and spatial distribution of vegetation and soil characteristics are key features for its survival, particularly in terms of feeding, locomotion, burrow construction and protection against predators. Importantly, C. nelsoni is a key ecological component of desert ecosystems, given its fundamental role for seed dispersal and soil removal, contributing to the ecosystem’s vegetation structure and dynamics. It is also an important component of the food chain as prey source of many reptiles, birds and carnivores (Patton 1970; Serrano 1987; Brown and Heske 1990a, 1990b).
The MBR is an area of great biological value due to the high species richness and endemicity it harbours (Serrano 1987; Grünberger 2004; CONANP Comisión Nacional de Áreas Naturales Protegidas (2006); WWF World Wildlife Fund (2019)). We have been studying the population and landscape genetics of the rodent species of the MBR for several years (Flores-Manzanero et al. 2019; Luna-Bárcenas 2023; unpublished data), building on ecological, genetic and evolutionary knowledge in a reserve that protects one of the best conserved desert ecosystems in North America. Considering the habitat features likely associated with C. nelsoni’s dispersal, distribution and survival, our objective in this study was to determine what landscape factors would influence its functional connectivity at a microgeographic scale in our study area within the MBR. To this end, we assessed the population genetic patterns of this non-model species (no reference genome is available for this or related species), using single-nucleotide polymorphisms (SNPs). We evaluated the relationship between landscape variables and gene flow under an isolation by resistance model. We predicted that soil type, configuration of vegetation and elevation, given their importance for the movement of individuals, building of burrows and feeding, would be key variables for dispersal (connectivity).
Materials and methods
Study site and sampling
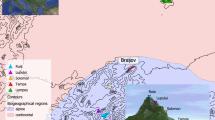
The Mapimí Biosphere Reserve is located in the Chihuahuan Desert of northern Mexico (26°35’—26°40’N—103°45’—104°00’W), with an elevation range of 135–1450 m, average temperature of 18.9 °C and annual precipitation of 242.8 mm (CONANP Comisión Nacional de Áreas Naturales Protegidas (2006)). Our study site covers an area of 15 km2 along the western slope of the Cerro San Ignacio (Fig. 1), characterized by xerophytic vegetation and shrubs (González 1983; CONANP Comisión Nacional de Áreas Naturales Protegidas (2006)). We sampled Nelson’s pocket mice during the breeding season (June 2015 and May 2017) with Sherman live traps (7.6 × 7.6 × 33 cm) baited with a mixture of rolled oaks, peanut butter and vanilla extract, during six consecutive nights. Trapping was performed along three transects per sampling night, separated 300 m and distributed following the presence of burrows to increase sampling success and to best cover the landscape heterogeneity (i.e. vegetation, soil, elevation; Montaña 1988). Each transect, of approximately 300 m length, consisted of up to 40 traps separated by ca. 5–10 m. Sampling intensity was 50–90 traps per night for a total of 840 traps. At each sampling site we recorded the vegetation species and geographic coordinates and took pictures of the soil surface for soil classification (see below). We sexed and took standard morphological measurements (Mills et al. 1995): body length (BL), tail length (T), hind foot length (FL), ear length (E) and weight (W); we also estimated the body mass index (BMI = W/BL2). A tissue sample (ear punch) was taken for subsequent genetic analysis and stored in labelled Eppendorf tubes with 96% ethanol. All individuals were released at the sampling site once data recording was finished, and individual well-being was assured. Procedures were conducted in strict accordance with the American Society of Mammalogists’ guidelines for use of wild mammal species (Sikes 2016) and with the corresponding collecting permits (FAUT 0168). No institutional ethical approval was required given no experimental procedures or killing of individuals were performed.
Sampling localities and spatial distribution of Chaetodipus nelsoni. Sampled individuals are indicated with blue dots and projected on a grayscale satellite picture of the study site (Rancho San Ignacio property). The darker grey depicts the San Ignacio Hill of the MBR. Photograph credit: Juan Cruzado (inaturalistaMX).
Morphological analyses
We evaluated the distribution of the six morphological measurements only in adult individuals (see “Results”) with the Shapiro-Wilk test (Royston 1982) with Stats v.3.4.3 (https://www.rdocumentation.org/packages/stats/versions/3.4.3) and performed multicollinearity tests with usdm v.1.1–18 (https://rdrr.io/rforge/usdm/f/). Because data were not normally distributed, we used Kruskal-Wallis tests (Hollander and Wolfe 1973) to evaluate differences between sexes and between the soil textures (described below in landscape data); the latter was also evaluated with a one-way Analysis of Variance (ANOVA) based on ranges (Hettmansperger and McKean 2011) with Rfit v.0.23 (Kloke and McKean 2012). We performed the same analyses to determine morphological differences between genetic groups (see Results). All statistical packages were run in R v.3.4.3 (R Core Team 2018).
Genotyping by sequencing and SNP calling
We extracted genomic DNA using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s instructions. To obtain an optimal sample quality for sequencing we homogenized samples into an average concentration of 50 ng/μl, measured using Qubit Fluorometric Quantification (Invitrogen, Carlsband, CA). We selected 95 samples based on DNA quality, maintaining an adequate representation of sampling distribution. Library preparation and sequencing was performed in the University of Wisconsin Biotechnology Centre (UWBC) following a Genotyping by sequencing approach (GBS; Elshire et al. 2011). Libraries were prepared using the ApeKI enzyme (cutting site: C[AT]G) and sequenced on a single lane of an Illumina Hi-Seq 2500 (Illumina Inc, USA) with single-end 101 bp reads.
A de novo assembly was performed using raw data processed with ipyrad (Eaton and Overcast 2020; https://ipyrad.readthedocs.io/en/master/), a simple and reproducible RADseq assembly and analysis framework that is computationally efficient. The assembly workflow in ipyrad is fully self-contained, capable of taking raw Illumina data and producing assembled output files without the need for pre- or post-processing by other software (Eaton and Overcast 2020). Considering that the parameters used in de novo locus identification and genotyping may affect downstream analyses and resulting inference, we examined a range of values of Mindepth_statistical, Clust_threshold, and min_samples_locus parameters to optimize assembly in the ipyrad pipeline (the rest of parameters were used as default). The final processing and filtering protocol used is described in Table S1.
Neutral genetic markers, which are not subject to natural selection, have great potential to empirically test functional connectivity, providing insights into population structure, gene flow and genetic diversity (Balkenhol et al. 2016). Hence, given the microgeographic scale of our study, the lack of a reference genome, and that the GBS sequencing yielded a high number of missing data, we focused on neutral loci for our main objectives of assessing the influence of landscape variables on gene flow and functional connectivity, and the potential association of genetics and morphology (Rellstab et al. 2016). We further processed our data using VCFtools v.0.1.13 (Danecek et al. 2011), testing different combinations of filtering parameters (--max-missing 0.5 and --mac 3; --max-missing 0.3 and --mac 3; --max-missing 0.2 and --mac 3; --max-missing and 0.2 --mac 2). The combination that included removing missing data per individual > 50% and -mac=3 rendered the best results, namely the highest number of samples and of SNPs, with lowest missing data and retaining one SNP per locus. To confirm the neutrality of the final loci obtained we applied BayeScan v.2.1, which uses allele frequencies to estimate the posterior probability of scenarios with or without selection, via a mixed Markov-chain model and Montecarlo simulations (Foll and Gaggiotti 2008). BayeScan was run by command line in macOS Sierra v.10.12.6 with the parameters: 50,000 iterations, 20 thinning intervals, 20 pilot runs with lengths of 10,000 iterations, 10,000 burn-in, and default settings for the remaining parameters. MCMC chain convergence was tested with Coda v.0.19–1 (Plummer et al. 2006) using a false discovery rate (FDR) < 0.05 and <0.01 threshold. This filtered final data set was used for all genomic analyses.
Genetic diversity and structure
We evaluated linkage disequilibrium of our SNP data with poppr v.2.8.1 in R (Kamvar et al. 2014). To estimate genetic variability, we calculated the allelic richness index, which uses rarefication to account for differences in sample sizes and genotyping success; the expected and observed heterozygosity and inbreeding coefficient (FIS) were estimated, all with PopGenReport v.3.0 in R (Adamack and Gruber 2014).
We inferred population structure with an array of analytical procedures. Two ordination methods were used, a principal component analysis (PCA) that considers genetic variation as a continuous value along orthogonal axes, with individuals grouped by similar ancestry (Ma and Amos 2012), using pcadapt v.4.0.3 in R (Luu et al. 2017); and a discriminant analysis of principal components (DAPC; Jombart 2008), an approach that does not assume any genetic model and uses the a-score method to determine the proportion of successful reassignment by individual in function of the number of retained PCs, with adegenet v.2.1.1 in R (Jombart 2008). We also used a Bayesian reconstruction of genealogical relationships to estimate structure and ancestry at the individual level, applying a sparse non-negative matrix factorization (sNMF) method (Frichot et al. 2014). It computes regularized least-square estimates of admixture proportion to estimate individual ancestry coefficients. This programme is well suited for SNPs and for datasets with missing data (Frichot et al. 2014). We ran sNMF with LEA v.2.6.0 in R (Frichot and François 2015) with 100,000 iterations, 50% burn-in, and 10 repetitions for K-values from 1 to 10; sNMF uses a cross-validation method in which a dataset with 10% of masked genotypes is created and a training set is used to evaluate the ability to correctly impute masked genotypes. Finally, as a complementary analysis we applied a spatially independent method based on a Bayesian inference of admixture proportions, performed with Structure v.2.3.4 (Pritchard et al. 2000) and using only the ipyrad filtered data set. We tested K = 1–5, did 10 independent runs with a burn-in of 10,000, followed by 100,000 MCMC iterations, assuming an admixture model and correlated allelic frequencies. The most likely number of genetic clusters (K) was assessed with the Evanno’s delta-K test with Structure-Harvester (Earl and vonHoldt 2012; web interface: http://taylor0.biology.ucla.edu/structureHarvester/). Results were summarized and averaged using CLUMPP v.1.1.2 (Jakobsson and Rosenberg 2007). We calculated two metrics to evaluate genetic differentiation between the genetic groups identified (see Results), FST and proportion of shared alleles (DPS; Bowcock et al. 1994) with hierfstat (Goudet 2005) and PopGenReport respectively, and also performed an analysis of molecular variance (AMOVA) with function poppr.amova in poppr.
We applied linear programming discriminant analysis (lpda in R; Nueda et al. 2022) to evaluate morphological differences between the genetic groups, performing one multivariate test (morphological measures were log10 transformed) and also testing the significant variable identified (hind foot length; see Results) alone. The lpda method looks for a hyperplane that separates the samples into two groups by minimizing the sum of all the distances to the subspace assigned to the group each individual belongs to (Nueda et al. 2022).
We performed Estimated Effective Migration Surfaces (EEMS; Petkova et al. 2016) to identify and visualize variation in gene flow, or effective migration rates, based on data deviation from the null expectation of isolation by distance under a stepping-stone model. EEMS uses georeferenced genetic data and migration patterns are fitted by the similarity between the genetic differences expected under the model and the genetic differences observed in the data. Estimates are then interpolated across the landscape to create the migration surface; if genetic similarity decreases rapidly in the observed data compared to the expected data, a low effective migration surface is inferred, namely a barrier to gene flow. We ran EEMS with runeems_snps, using a deme size of 200, with three independent starting chains for 5 × 106 MCMC iterations, 5 × 104 of burn-in, thinning of 5000 and different starting seeds. Convergence of runs was confirmed with the eems.plots function, which were then combined and visualized, using reemsplots2 in R (https://github.com/dipetkov/reemsplots2).
Landscape data
We chose four landscape variables potentially associated with individual movement and dispersal of C. nelsoni (Serrano 1987; Best 1994; Ceballos and Oliva 2005): elevation, vegetation cover (as the normalized difference vegetation index; NDVI), physiography (classification from Montaña 1988) and soil texture. Each variable was represented as a raster layer, encompassing our study area (4 × 3 km) at a fine spatial scale (15 m). Elevation values were obtained from the ‘Mexican elevational continuum surfaces’ v3.0 (resolution of 15 m, available at https://www.inegi.org.mx/app/geo2/elevacionesmex/); values are continuous and in metres. We calculated the NDVI index using ERDAS Imagine v.13.0 and ArcMap v.10.2.1 from a Landsat 8 image (ID: LC80250472019086LGN00; downloaded on 15 June 2017 from the Earth explorer platform; https://earthexplorer.usgs.gov), to obtain a raster surface with continuous values. NDVI has values from −1 to 1, based on the behaviour of vegetation and soils in the red and near‐infrared spectral regions (Silleos et al. 2006); areas of sand or barren rock show low values (0.1 or less), sparse vegetation (e.g. grasslands, shrubs) ca. 0.2–0.5. and high vegetation cover closer to 1.0. Physiography describes the general aspects of geology, geomorphology and soils of a region. Based on the physiographic classification of Montaña (1988) for the Mapimí Biosphere Reserve, we performed a supervised classification on the same Landsat image to generate the physiography surface. Briefly, we used the Signature editor in ERDAS to capture the spectral signatures of each physiographic classification and we draw polygons (training sets) across the area that encompassed each class. The corresponding spectral signature values were averaged to generate the supervised final raster layer that included five categorical values (Sb, Fb2, Bpg, Bg3, Sa; see Table S2 for definitions). We also performed a supervised classification for soil texture, based on our field observations and pictures from each sampling site, classified in three categorical values (Rocky, RockSand, Sandy; see Table S2 for definitions and Fig. S1).
We were able to represent the landscape heterogeneity of our study area as evaluated with the raster layers (Fig. 2). The elevation layer exhibited a range of 1150 to 1450 m (Fig. 2a), the NDVI index showed low values (0.05–0.15) indicating low vegetation cover, with spread bare soil areas mixed with shrubs and desert vegetation (Fig. 2b), the physiographic classification identified the Sb, Sa and Fb2 classes for the higher elevation sites and the Bpg and Bg3 for the lower zones (Fig. 2c); lastly, the soil texture layer showed the higher zone predominantly with rocks (Rocky type), following a gradient to RockSand to mostly sandy soil in the lower elevation (Fig. 2d).
Layers used for the resistance analyses, encompassing our study area (4 × 3 km) at our fine spatial scale (15 m). The blue dots indicate the geographic position of the sampled individuals. Continuous variables layers (a) elevation and (b) vegetation cover (as the normalized difference vegetation index; NDVI); categorical variables (c) physiography (classification from Montaña 1988) and (d) soil texture. See Table S2 for variables definitions and Fig. S1.
Landscape genetics and functional connectivity
We estimated geographic Euclidean distances with gstudio v.1.5.0 (Dyer 2014) in R, while genetic distance between pairs of individuals was estimated based on the proportion of shared alleles (DPS) in adegenet; DPS is an adequate metric because it does not make biological assumptions and has proved best for obtaining accurate inferences in landscape genetics analyses at the individual level (Bowcock et al. 1994; Shirk et al. 2017) and fine landscape scales (Flores‐Manzanero et al. 2019). We performed landscape genetic analyses to assess the effect of the landscape variables on C. nelsoni’s functional connectivity (see the predictions tested in Table S3) by applying the optimization framework developed by Peterman and collaborators (2014) with ResistanceGA v.4.0 in R (Peterman 2018; https://github.com/wpeterman/ResistanceGA), to determine the resistance values of our environmental surfaces with no a priori assumptions about the scale and direction of the resistance relationship. This approach utilizes a genetic algorithm (GA; Scrucca 2013) that adaptively explores the parameter space of monomolecular and Ricker functions (Bolker 2008) to transform continuous surfaces into resistance surfaces. In the process, the GA seeks to maximize the relationship between pairwise genetic and landscape effective distances. It has the advantage that raster layers of both continuous and categorical variables can be used. It performs a model selection procedure, an optimization of combined surfaces (multivariate model), and inferences about the contribution of each surface to total resistance in multivariate models (Cushman et al. 2006; Peterman 2018).
We optimized the resistance values of categorical (physiographic classification and soil texture) surfaces prior to applying the optimization framework, to represent landscape features as continuously distributed, which are more ecologically relevant when handling categorical or binary surfaces (Peterman 2018). We used the k.smooth function in ResistanceGA. Next, all surfaces were independently optimized estimating pairwise effective distances among individuals using the commuteDistance function with gdistance in R (Van Etten (2017)), which is functionally equivalent to Circuitscape (Kivimäki et al. 2014; Peterman 2018) but is computationally faster and can be run in parallel. We fit linear mixed-effects models for each competing landscape surface (hereafter “model”) by considering the non-independence inherent in pairwise distance matrices, via a maximum-likelihood population effects (MLPE) parameterization (Clarke et al. 2002; Van Strien et al. 2012) using lme4 in R (Bates et al. 2014). The response variable was the genetic distance, and the predictor variables were the scaled and centred landscape cost distances, including the Euclidean distance model. We used the AICc (Akaike’s information criterion corrected for small/finite sample size; Akaike 1974) obtained from the fitted MLPE model to establish which model was related to the individual-based genetic differentiation (indicated by the lowest AIC score).
Given that environmental variables can affect individuals synergistically, we also performed multivariate models with ResistanceGA. We performed a bootstrap resampling of the data to evaluate the robustness of the model given different combinations of samples (Peterman et al. 2014; Ruiz-Lopez et al. 2016). For this, 75% of the samples were randomly selected without replacement and then each surface was fit to the subset of samples; the average rank, average model weight, and the percentage that a surface was selected as the best model were estimated following 10,000 iterations with Resist.boot function. Finally, we performed multi-surface optimization for the composite surfaces (hereafter, multivariate models), and performed again a bootstrap model selection with the same previous parameters to obtain the average rank, average model weight, and the selection percentage of both univariate and multivariate surfaces (Peterman et al. 2014; Peterman 2018). Importantly, we also performed this entire sequential approach to analyse the resistance patterns between the two genetic clusters obtained (see Results).
Results
Sampling and morphology
We sampled a total of 133 Chaetodipus nelsoni individuals (we did not consider pregnant and lactating females), including 73 adults (41 males, 32 females), 38 juveniles and 22 undetermined (Table S4). We documented six species of plants associated with the burrows where (or closest to) individuals were trapped: Agave sp., Euphorbia antisyphilitica, Jatropha dioica, Prosopis glandulosa, Opuntia sp. and Larrea tridentata; the latter was found with higher frequency in the sampling sites with highest trapping success (Fig. S1d).
The morphological analyses were based on the measurements of the 73 adult individuals (Table S5), none of which had a normal distribution (Shapiro-Wilk), while the body mass index showed collinearity and was excluded. There were no statistical differences between males and females nor among the different soil textures (Kruskal-Wallis). On the other hand, the ANOVA results showed significant differences for the hind foot (FL) and the type of soil (Fig. 3).
Box plots for the comparison between five morphological measurements of 74 adult Chaetodipus nelsoni individuals and the three soil texture classes (1: Rocky, 2: RockSand, 3: Sandy; see Table S2). The hind foot length showed significant differences (ANOVA).
Genetic diversity and structure
Results from GBS rendered a total of 210,734,772 raw reads, an average number of raw reads per sample of 5.48x105 and average coverage = 6x. The ipyrad and VCFtools filtering rendered 718 SNPs for 73 individuals. BayeScan results indicated that none of the 718 loci had significant selection signal (FDR > 0.05; Fig. S2a). Finally, for the building of the landscape raster layers, we selected one individual per pixel (to avoid duplicated data) while keeping representation of the entire study area, thus our final data set included 67 individuals and 718 SNPs. Genetic results did not show linkage disequilibrium (Fig. S2b), while diversity values were allelic richness = 1.8, observed heterozygosity = 0.031 and significantly different expected heterozygosity = 0.115 (p < 0.001). We did not detect inbreeding signals (FIS = −0.085 and −0.096, High and Low elevation groups respectively).
All structuring analyses rendered consistent results, grouping the same individuals: principal component (PCA; Fig. 4a) and discriminant analysis of principal components (DAPC; Fig. S3a) showed data separated in two distinct genetic groups, one corresponding with individuals from the higher elevation (High elevation) and the other from the lower elevation (Low elevation) sampling sites of the Cerro San Ignacio (Fig. 4b). sNMF results also identified K = 2 as the optimum number of genetic clusters (Fig. 4c), followed by K = 3 (Fig. S3b); concurringly, Structure showed K = 2 with highest likelihood (Fig. S3c). AMOVA results indicated that differentiation between and within genetic groups explained 92.5% (sigma = 8.4708) and 7.4% (sigma = 0.681) of the variation (p = 0.01), respectively; genetic indices also showed high differentiation (FST = 0.4187; DPS = 0.2865) between groups. Finally, discriminant analysis results for the morphological multivariate test showed a 74–84% genetic group prediction (Fig. S4a), while it was 100% prediction of individuals per group with the hind foot variable (Fig. S4b). The EEMS results exhibited regions of lower than expected migration in accordance with the two groups; specifically, regions of significantly reduced or null gene flow (orange colour in Fig. 4d) between the two groups, and high migration (blue colour) within each group. Finally, this consistent genetic separation between groups is complemented with the significant (p < 0.001) morphological difference observed for the hind foot length (FL) associated with the soil texture (Fig. 4e). That is, the individuals from High elevation had an average FL of 19.5 mm associated with the Rocky soil from the higher elevational areas, whereas individuals from Low elevation showed an average FL of 21 mm associated with the Sandy soil and the lower elevation area.
a Results of the principal components analysis (PCA) depicting two distinct genetic groups (High elevation and Low elevation); axis 1 and axis 2 explain 8% and 6% of the variance, respectively. b Satellite image showing the geographic location of the sampled individuals, indicating the two genetic groups identified: High elevation in green and Low elevation in red circles. c The two groups identified with the sNMF analysis of ancestry proportion; the bar plot indicates individual membership (each vertical line depicts one individual). d Migration patterns (EEMS results) depicting regions of significantly reduced or null gene flow (in orange) and high migration (in blue). e Box plot of the comparison between the hind foot length of the two genetic groups individuals.
Landscape genetics and functional connectivity patterns
In ResistanceGA, each variable was considered as a model to evaluate the observed genetic structure, while comparing with a geographic (Euclidian distance) and a null model as well. Results of model selection from the optimization of univariate models showed that all four variables (NDVI, elevation, physiographic classification and soil texture) were better supported models in comparison with the distance and null models, significantly explaining (t > 1.96) the genetic differentiation (genetic distance DPS) (Table S6). Given that all univariate models exhibited a high selection, we used the four the variables to perform multi-surface optimization. Model selection showed that the combination of physiographic classification and soil texture was the best-supported (Table 1).
Results of the resistance optimized models regarding the two genetic groups showed a distinct resistance pattern for each group (Fig. 5, Table S7). Namely, the elevation model exhibited an Inverse Ricker function, assigning high resistance to areas below 1200 m and the lowest resistance between 1250 and 1350 m for High elevation (Fig. 5a, e). Comparatively, in Low elevation an Inverse Reverse monomolecular functional form was observed, where resistance increases with elevation (i.e. the lowest resistance is at the lowest elevation; Fig. 5a, f). Also, an Inverse Ricker function was observed for the NDVI model in High elevation, which assigned high resistance to areas lacking vegetation (< 0.05), low resistance where it is scarce (>0.05 < 0.1) and increasing towards areas with more vegetation present (>0.1) (Fig. 5b, g). This pattern is different in Low elevation, in which the function was Inverse-reverse Ricker, with lower resistance where sandy soils and shrub vegetation predominate (0.15) (Fig. 5b, h). The Bpg and Fb2 physiographic classes (gravel, basalt and volcanic tuff) exhibited lower resistance for High elevation, while for Low elevation were Bpg and Bg3 (footslopes with gravel sediments) (Fig. 5c, Table S7). Finally, patterns were also contrasting regarding soil texture: lowest and highest resistance was observed for the Rocky and the Sandy soils for High elevation and Low elevation, respectively (Fig. 5d, Table S7).
Best supported resistance optimized models for four landscape variables, showing different resistance patterns between the two genetic groups identified. The lighter colours in the scale of grey depict areas with lower resistance, while the darker ones indicate higher resistance. The green and red circles correspond to High elevation and Low elevation genetic groups, respectively. Continuous variables layers (a) elevation (m) and (b) vegetation cover (as the normalized difference vegetation index; NDVI); categorical variables (c) physiography (classification from Montaña 1988) and (d) soil texture. e–h Elevation and NDVI models: High elevation showed an Inverse Ricker function for both (e) Elevation, assigning high resistance to areas below 1200 m and the lowest resistance between 1250 and 1350 m; and (g) NDVI with high resistance to areas lacking vegetation (<0.05), low resistance where it is scarce (>0.05 < 0.1), and increasing in areas with more vegetation (>0.1). f Low elevation had an Inverse Reverse monomolecular function for Elevation, where resistance increases with elevation (i.e. the lowest resistance is at the lowest elevation); and (h) NDVI showed and Inverse-reverse Ricker, with lower resistance where sandy soils and shrub vegetation predominate (0.15).
Discussion
Microgeographic genetic structuring of Nelson’s pocket mouse
In terms of genetic diversity, C. nelsoni showed moderate allelic diversity and heterozygosity, similar to values present in Dipodomys rodents in the study region (Busch et al. 2007; Flores-Manzanero et al. 2019), whereas no comparable genomic surveys have been performed with this or other Chaetodipus species. Despite the microgeographic spatial scale of our study, our findings show that C. nelsoni is clearly differentiated in two genetic clusters, significantly associated with the heterogeneity of the landscape. The robustness of the genetic group structure is evidenced by the different methods used (PCA, DAPC, sNMF, EEMS) and the genetic differentiation observed (FST = 0.4187; DPS = 0.2865; AMOVA) (Shirk et al. 2017). Two rodent species, Dipodomys nelsoni and D. merriami from the same study area and scale show little to null genetic structuring, respectively (Flores-Manzanero et al. 2019; Luna-Bárcenas 2023).
Finding genetic differences at a fine (micro) scale is challenging because it depends, among others, on the biological features of the organisms studied (e.g. vagility, phylopatry, behaviour) and the environmental heterogeneity of their habitat (Anderson et al. 2010; Reding et al. 2013; Russo et al. 2016). Neutral population genetic structure entails allele frequency differences among populations that have arisen due to neutral processes such as genetic drift, gene flow and mutation. Given our aim of determining, at a markedly microgeographic scale and at the individual level, the landscape variables that influence C. nelsoni spatial genetic structure and gene flow, we focused on neutral diversity (Balkenhol et al. 2016; Rellstab et al. 2016). In view of our rigorous filtering and SNP selection, we are confident in our genomic data (718 SNPs). This number of loci can result from an intrinsically low genetic diversity in the population and/or due to the small area studied that does not capture the species overall variability. Dipodomys nelsoni from the same study site shows a similar pattern of low number of loci and moderate genetic structure (Luna-Bárcenas 2023).
Along with C. nelsoni genetic differentiation in two groups, we found that dispersal (effective migration) is significantly limited along the region separating the two groups while it is high within each group’s distribution (Fig. 4), suggesting potential barriers to gene flow and low individual movement, while their requirements (e.g. food, shelter, reproduction) are likely fulfilled at such fine scale. Although C. nelsoni’s social system is unknown, the fact that it builds similar burrow networks as desert Dipodomys rodents (Best et al. 1988) and that adult males disperse during the reproductive season (Aguilera-Miller et al. 2018), suggest C. nelsoni is organized in small family groups (Best 1994). Our sampling concurs with this, as we consistently observed/captured few individuals per burrow. Male-biased dispersal contributes to avoid inbreeding in this kind of family groups where related individuals share burrows, like in Dipodomys nelsoni, D. spectabilis and other rodents like squirrels, marmots and prairie dogs (Schwartz and Armitage 1980; VanStaaden 1994; Dobson 1998; Hoogland 2002), which is supported by the lack of inbreeding indicated by our results.
Landscape features influencing Chaetodipus nelsoni’s functional connectivity
Dispersal is a fundamental life history aspect that enables individuals to maximize fitness by optimizing resource access, including food and reproductive partners, while reducing predation risk, all of which is in turn influenced by the environment (Ronce 2007; Manel and Holderegger 2013; Reding et al. 2013; Garrido-Garduño et al. 2022). As we predicted, elevation, vegetation and soil were key environmental variables for the connectivity of C. nelsoni while, interestingly, patterns differed between genetic groups. Low resistance patterns are indicative of dispersal, i.e. that connectivity is facilitated, as shown by our migration results denoting high gene flow (connectivity) within and low gene flow between C. nelsoni genetic groups.
Elevation is a key ecological factor for C. nelsoni since it is linked with vegetation gradients (Geluso and Geluso 2015) and soil physiography across its habitat in the MBR (Montaña 1988). Accordingly, results show that one genetic group (High elevation) has lower resistance on the more elevated zones of Cerro San Ignacio (1250–1350 m) in areas with scarce and scattered vegetation, while connectivity in Low elevation group is facilitated in the lower (< 1200 m), more flat areas and with more vegetation cover. Vegetation, measured as NDVI, was also significantly associated with functional connectivity; this metric has proven to adequately capture vegetation heterogeneity in Dipodomys (Flores-Manzanero et al. 2019; Luna-Bárcenas 2023), as well as in other taxa (e.g. birds and mammals, Borja-Martínez et al. 2022; White et al. 2022; lizards, Romero-Báez et al. 2024; amphibians, Gutiérrez-Rodríguez et al. 2017). Specificity to shrub vegetation in desert-dwelling organisms has been documented as a key feature in rodents for feeding (seeds) and shelter (burrows) (Matson and Baker 1986; Best 1994; Flores-Manzanero et al. 2019; Luna-Bárcenas 2023), as well as in other species like the endangered lizard Gambelia sila (Westphal et al. 2018). Chaetodipus nelsoni is known to build burrows at the bases of certain plant species like Astrolepis sp., Fouquieria splendens, Echinocereus, Agave lechuguilla, Acacia greggii, A. roemeriana, Mimosa biuncifera, Viguiera stenoloba, Dasylirion sp., Parthenium incanum (Serrano 1987; Geluso and Geluso 2015). Interestingly, we found that in the MBR it builds burrows associated with two not previously reported plant species, Euphorbia antisyphilitica and Jatropha dioica.
Soil type and soil texture are also fundamental factors for desert rodents like Chaetodipus and Dipodomys species, associated with the fact that they build burrows and with their kind of locomotion (Hall et al. 2022), bipedal in kangaroo rats and both hopping and moving on all four feet in C. nelsoni (Randall 1993; Best 1994). Furthermore, size of rocks is known to be of great importance in determining the abundance of C. nelsoni (Best 1994), while the architecture and dimensions of burrows vary depending on the type of soil in desert and other rodent species (Best et al. 1988; Kocher, Parshad (2003)). Accordingly, High elevation has higher connectivity in the elevated zones with presence of basalt and turf and where the surface is dominated by medium-large rocks, and contrastingly Low elevation does where gravel and sandy soils predominate.
Notably, we also identified a morphological characteristic that is significantly different between genetic groups, the hind foot length, where individuals from Low elevation have larger feet (21 mm) than those in High elevation (19.5 mm). We suggest it is associated with the soil characteristics and landscape features (elevation, vegetation), since locomotion types (hopping, running) and excavating for building burrows depend on hind feet size and strength (Verde-Arregoitia et al. 2017). In summary, we describe novel morphological, ecological and genetic aspects of the Nelson’s pocket mouse C. nelsoni in the Mapimí Biosphere Reserve (Chihuahuan desert). We show it is microgeographically structured in two genetic groups and its functional connectivity is clearly influenced by landscape variables that are ecologically relevant and associated with morphology, behaviour and life history traits (Fig. 6). Further genomic and morphometric studies are needed to unravel if potential local adaptation processes are taking place, associated with two ecomorphs with distinct habitat preferences. They should be designed to explicitly assess adaptive loci and phenotype- and genome-wide-associations. Finally, the patterns observed improve our understanding of the genetic, ecological and behavioural aspects of C. nelsoni, as well as of the ecology and life history of desert rodents.
Representation of the functional connectivity of each genetic group: the elevational gradient (1150–1450 m) is depicted as a triangle, circles represent the soil texture (sandy at the lower and rocky at the higher elevations), while vegetation cover is shown with cacti silhouettes, scattered at the top and with higher vegetation cover at the base. Rodent genetic groups and significant size difference of the hind foot length are shown (High elevation in green, Low elevation in red). The grey scale on the sides of the triangle represents the resistance levels associated with the landscape variables, in which darker grey depicts where connectivity is lower (highest resistance) and lighter grey indicates higher connectivity (lowest resistance). Photo: C. nelsoni at the entrance of its burrow in the Mapimí Biosphere Reserve. (Photo credit: Carlos Luna-Aranguré).
Data availability
Input files for population genetic analyses (raw and filtered genomic data files) have been made available on Dryad (https://doi.org/10.5061/dryad.qbzkh18sz).
References
Adamack AT, Gruber B (2014) PopGenReport: Simplifying basic population genetic analyses in R. Methods Ecol Evol 5:384–387
Aguilera-Miller E, Álvarez-Castañeda ST, Murphy R (2018) Matrilineal genealogies suggest a very low dispersal in desert rodent females. J Arid Environ 152:28–36
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Anderson CD, Bryan KE, Marie JF, Holderegger R, James PM, Rosenberg MS et al. (2010) Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol Ecol 19:3565–3575
Bates DM, Maechler M, Bolker BM, Walker S (2014) Linear mixed-effects models using Eigen and S4. R package version 1.1-6. Retrieved from http://CRAN.R-project.org/package=lme4
Balkenhol N, Cushman S, Storfer A, Waits L (2016) Landscape genetics: Concepts, methods and applications. John Wiley & Sons Ltd, Chichester, UK
Best TL (1994) Chaetodipus nelsoni. Mamm Species 484:1–6
Best TL, Intress C, Shull KD (1988) Mound structure in three taxa of Mexican kangaroo rats (Dipodomys spectabilis cratodon, D. s. zygomaticus and D. nelsoni). Am Midl Naturalist 119:216–220
Bolker B (2008) Ecological models and data in R. Princeton University Press, Princeton
Borja-Martínez G, Tapia-Flores D, Shafer ABA, Vázquez-Domínguez E (2022) Highland forest’s environmental complexity drives landscape genomics and connectivity of the rodent Peromyscus melanotis. Landsc Ecol 37:1653–1671
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza L (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457
Brown JH, Heske EJ (1990a) Temporal changes in a Chihuahuan desert rodent community. Oikos 59:290–302
Brown JH, Heske EJ (1990b) Control of a desert‐grassland transition by a keystone rodent guild. Science 250:1705–1707
Busch JD, Waser PM, DeWoody JA (2007) Recent demographic bottlenecks are not accompanied by a genetic signature in banner-tailed kangaroo rats (Dipodomys spectabilis). Mol Ecol 16:2450–2462
Castillo JA, Epps CW, Davis AR, Cushman SA (2014) Landscape effects on gene flow for a climate-sensitive montane species, the American pika. Mol Ecol 23:843–856
Ceballos G Oliva G (2005) Los mamíferos silvestres de México. Conabio/Fondo de Cultura Económica, México
Clarke RT, Rothery P, Raybould AF (2002) Confidence limits for regression relationships between distance matrices: Estimating gene flow with distance. J Agric Biol Environ Stat 7:361–372
CONANP (Comisión Nacional de Áreas Naturales Protegidas) (2006) Programa de conservación y manejo, Reserva de La Biosfera Mapimí. Conanp, México
Cosentino BJ, Schooley RL, Bestelmeyer BT, McCarthy AJ, Sierzega K (2015) Rapid genetic restoration of a keystone species exhibiting delayed demographic response. Mol Ecol 24:6120–6133
Cushman SA, McKelvey KS, Hayden J, Schwartz MK (2006) Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Naturalist 168:486–499
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, Depristo MA et al. (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Dobson FS (1998) Social structure and gene dynamics in mammals. J Mamm 79:667–670
Dyer RJ (2014) gstudio. An R package for the spatial analysis of population genetic data. https://github.com/dyerlab/gstudio
Earl DA, vonHoldt BM (2012) Structure Harvester: A website and program for visualizing structure output and implementing the evanno method. Conserv Genet Res 4:1–3
Eaton DAR, Overcast I (2020) ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 36:2592–2594
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple Genotyping-by-Sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Ann Rev Ecol 34:487–515
Flores-Manzanero A, Vázquez-Domínguez E (2019) Landscape genetics of mammals in American ecosystems. Therya 10:381–393
Flores-Manzanero A, Luna-Bárcenas MA, Dyer RJ, Vázquez-Domínguez E (2019) Functional connectivity and home range inferred at a microgeographic landscape genetics scale in a desert‐dwelling rodent. Ecol Evol 9:437–453
Frankham R (2010) Challenges and opportunities of genetic approaches to biological conservation. Biol Conserv 143:1919–1927
Frichot E, François O (2015) LEA: An R package for landscape and ecological association studies. Methods Ecol Evol 6:925–929
Frichot E, Mathieu F, Trouillon T, Bouchard G, François O (2014) Fast and efficient estimation of individual ancestry coefficients. Genetics 4:973–983
Foll M, Gaggiotti OE (2008) Genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Garrido-Garduño T, Téllez-Valdés O, Manel S, Vázquez-Domínguez E (2016) Role of habitat heterogeneity and landscape connectivity in shaping gene flow and spatial population structure of a dominant rodent species in a tropical dry forest. J Zool 298:293–302
Garrido-Garduño T, Vázquez-Domínguez E, Dávila-Aranda P, Lira-Saade R, Arenas-Navarro M, Téllez-Valdés O (2022) Defining environmentally heterogeneous sites when faced with conservation urgency and scarce in situ data. Rev Mex Biodiv 93:e934132
Geluso KN, Geluso K (2015) Distribution and natural history of Nelson’s pocket mouse (Chaetodipus nelsoni) in the Guadalupe mountains in southeastern New Mexico. Occasional Pap Mus Tex Tech Univ 332:1–20
González ES (1983) La Vegetación de Durango. Serie: Cuadernos de Investigación Tecnológica, Durango, México
Goudet J (2005) Hierfstat, a package for R to compute and test variance components and F-statistics. Mol Ecol Notes 5:184–186
Grünberger O (2004) Características esenciales de la Reserva de la Biosfera. In: Grünberger O, Reyes-Gómez V.M, Janeau JL (eds) Las playas del desierto chihuahuense (parte mexicana): Influencia de las sales en ambiente árido y semiárido. Institut de Recherche pour le Développement. Instituto de Ecología, A.C., México, pp 41–55
Gutiérrez-Rodríguez J, Gonçalves J, Civantos E, Martínez-Solano I (2017) Comparative landscape genetics of pond-breeding amphibians in Mediterranean temporal wetlands: The positive role of structural heterogeneity in promoting gene flow. Mol Ecol 26:5407–5420
Hall JK, McGowan CP, Lin DC (2022) Comparison between the kinematics for kangaroo rat hopping on a solid versus sand surface. R Soc Open Sci 9:211491
Hettmansperger TP, McKean JW (2011) Robust nonparametric statistical methods. 2nd edition. CRC press, Florida, USA
Hoban S, Bruford M, D’urban JJ, Lopes-Fernandes M, Heuertz M, Hohenlohe PA et al. (2020) Genetic diversity targets and indicators in the CBD post-2020 Global Biodiversity Framework must be improved. Biol Conserv 248:108654
Hollander M, Wolfe DA (1973) Nonparametric statistical methods. John Wiley & Sons, Inc, USA
Hoogland JL (2002) Sexual dimorphism of prairie dogs. J Mamm 84:1254–1266
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T (2008) Adegenet: An R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 4:281
Kivimäki I, Shimbo M, Saerens M (2014) Developments in the theory of randomized shortest paths with a comparison of graph node distances. Phys A: Stat Mech its Appl 393:600–616
Kloke JD, McKean JW (2012) Rfit: Rank-based estimation for linear models. R J 4:57–64
Kocher D, Parshad VR (2003) Structural and functional analysis of burrows of three rodent species in wheat fields in sandy and loamy soils in Punjab (India). Mammalia 67:603–605
LaPoint S, Balkenhol N, Hale J, Sadler J, Van der Ree R (2015) Ecological connectivity research in urban areas. Funct Ecol 29:868–878
Luna-Bárcenas MA (2023) Asociación de loci con factores ambientales en la estructura genómica de Dipodomys nelsoni. Dissertation, Instituto de Ecología, UNAM
Luu K, Bazin E, Blum GB (2017) Pcadapt: An R package to perform genome scans for selection based on principal component analysis. Mol Ecol Res 17:67–77
Ma J, Amos CI (2012) Principal components analysis of population admixture. PLoS ONE 7:e40115
Manel S, Holderegger R (2013) Ten years of landscape genetics. Trends Ecol Evol 28:614–621
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197
Marrotte RR, Gonzalez A, Millien V (2014) Landscape resistance and habitat combine to provide an optimal model of genetic structure and connectivity at the range margin of a small mammal. Mol Ecol 23:3983–3998
Matson JO, Baker RH (1986) Mammals of Zacatecas. Spec Publ Mus, Tex Tech Univ 24:1–88
Mills J, Yates TL, Childs JE, Parmenter RR, Ksiazek TG, Rollin PE, Peters CJ (1995) Guidelines for working with rodents potentially infected with hantavirus. J Mamm 76:716–722
Mimura M, Yakahara T, Faith DP, Vázquez-Domínguez E, Colautti RI, Araki H et al. (2017) Understanding and monitoring the consequences of human impacts on intraspecific variation. Evol Appl 10:121–139
Modi WS (2003) Morphological, chromosomal, and molecular evolution are uncoupled in pocket mice. Cytogenet Genome Res 103:150–154
Montaña C (1988) Estudio integrado de los recursos de vegetación, suelo y agua en la Reserva de La Biósfera de Mapimí. Instituto de Ecología, A.C., México
Mullins J, Ascensão F, Simões L, Andrade L, Santos-Reis M, Fernandes C (2015) Evaluating connectivity between Natura 2000 sites within the montado agroforestry system: a case study using landscape genetics of the wood mouse (Apodemus sylvaticus). Landsc Ecol 30:609–623
Neiswenter SA, Hafner DJ, Light JE, Cepeda GD, Kinzer KC, Alexander LF, Riddle BR (2019) Phylogeography and taxonomic revision of Nelson’s pocket mouse (Chaetodipus nelsoni). J Mamm 100:1847–1864
Nueda MJ, Gandía C, Molina MD (2022) LPDA: A new classification method based on linear programming. PloS ONE 17(7):e0270403
Patton JL (1970) Karyotypes of five species of pocket mouse, Perognathus (Rodentia: Heteromyidae), and a summary of chromosome data for the genus. Mamm Chromosome Newsl 11:3–8
Patton JL (2005) Family Heteromyidae. In: Wilson DE, Reeder DM (eds) Mammal species of the World: A taxonomic and geographic reference, 3rd edition. Johns Hopkins University Press, USA, pp 844-858
Patton JL, Sherwood SW, Yang SY (1981) Biochemical systematics of chaetodipine pocket mice, genus Perognathus. J Mamm 62:477–492
Peterman WE (2018) ResistanceGA: An R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol Evol 9:1638–1647
Peterman WE, Connette GM, Semlitsch RD, Eggert LS (2014) Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol Ecol 23:2402–2413
Petkova D, Novembre J, Stephens M (2016) Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet 48:94–100
Plummer M, Best N, Cowles K, Vines K (2006) CODA: Convergence diagnosis and output analysis for MCMC. R N. 6:7–11
Primack RB (2014) Essentials of conservation biology. Sinauer Associates, Sunderland
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2018) R: A language and environment for statistical computing. R foundation for statistical computing. https://doi.org/10.1007/978-3-540-74686-7
Randall JA (1993) Behavioural adaptations of desert rodents (Heteromyidae). Anim Behav 45:263–287
Reding DM, Cushman SA, Gosselink TE, Clark WR (2013) Linking movement behavior and fine-scale genetic structure to model landscape connectivity for bobcats (Lynx rufus). Landsc Ecol 28:471–486
Rellstab C, Zoller S, Walthert L, Lesur I, Pluess AR, Graf R et al. (2016) Signatures of local adaptation in candidate genes of oaks (Quercus spp.) with respect to present and future climatic conditions. Mol Ecol 25:5907–5924
Romero-Báez Ó, Murphy M, Díaz de la Vega-Pérez AH, Vázquez-Domínguez E (2024) Environmental and anthropogenic factors mediating the functional connectivity of the mesquite lizard along the eastern Trans-Mexican Volcanic Belt. Mol Ecol 33:e17469
Ronce O (2007) How does it feel to be like a rolling stone? Ten questions about dispersal evolution. Annu Rev Eco Evol Syst 38:231–253
Royston JP (1982) An extension of Shapiro and Wilk’s w tests for normality to large samples. Appl Stat 31:115–124
Ruiz‐Lopez MJ, Barell C, Rovero F, Roos C, Peterman WE, Ting N (2016) A novel landscape genetic approach demonstrates the effects of human disturbance on the Udzungwa red colobus monkey (Procolobus gordonorum). Heredity 116:167–176
Russo IR, Sole CL, Barbato M, Von Bramann U, Bruford MW (2016) Landscape determinants of fine-scale genetic structure of a small rodent in a heterogeneous landscape (Hluhluwe-IMfolozi Park, South Africa). Sc Rep. 6(29168):1–14
Schoville S, Bonin A, Francois O, Lobreaux S, Melodelima C, Manel S (2012) Adaptive genetic variation on the landscape: methods and cases. Annu Rev Ecol Syst 43:23–43
Schwartz A, Armitage KB (1980) Genetic variation in social mammals: the marmot model. Science 207:665–667
Scrucca L (2013) GA: A package for genetic algorithms in R. J Stat Softw 53:1–37
Serrano V (1987) Las comunidades de roedores desertícolas del Bolsón de Mapimí. Acta Zool Mex 1:1–22
Shirk AJ, Landguth EL, Cushman SA (2017) A comparison of individual-based genetic distance metrics for landscape genetics. Mol Ecol Res 17:1308–1317
Silleos G, Thomas A, Ioannis G, Konstantinos K (2006) Vegetation indices: Advances made in biomass estimation and vegetation monitoring in the last 30 years. Geocarto Int 21:21–28
Sikes RS (2016) Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J Mamm 97:663–688
Spear SF, Cushman SA, McRae H (2015) Resistance surface modeling in landscape genetics. In: Balkenhol N, Cushman SA, Storfer AT, Waits LP (eds) Landscape Genetics: Concepts, Methods, Applications. John Wiley & Sons Ltd, Chichester, UK, pp. 129-148
Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP (2010) Landscape genetics: Where are we now? Mol Ecol 19:3496–3514
Van Etten J (2017) R package Gdistance: Distances and routes on geo-graphical grids. J Stat Softw 76:1–21
Van Strien MJ, Keller D, Holderegger R (2012) A new analytical approach to landscape genetic modelling: Least‐cost transect analysis and linear mixed models. Mol Ecol 21:4010–4023
VanStaaden JM (1994) Suricata suricatta. Mamm Species 483:1–8
Vaughan TA, Ryan JA, Czaplewski NJ (2000) Mammalogy, fourth ed. Harcourt College Publishers, USA
Verde-Arregoitia LD, Fisher DO, Schweizer M (2017) Morphology captures diet and locomotor types in rodents. R Soc Open Sci 4:2054–5703
Wang IJ, Bradburd GS (2014) Isolation by environment. Mol Ecol 23:5649–5662
White JG, Sparrius J, Robinson T, Hale S, Lupone L, Healey T et al. (2022) Can NDVI identify drought refugia for mammals and birds in mesic landscapes? Sci Total Environ 851:158318
Whitford W (2002) Ecology of desert systems. Academic Press, California
Westphal MF, Noble T, Butterfield HS, Lortie CJ (2018) A test of desert shrub facilitation via radiotelemetric monitoring of a diurnal lizard. Ecol Evol 8:12153–12162
WWF (World Wildlife Fund) (2019) Deserts and xeric shrublands. Recovered from https://www.worldwildlife.org/biomes/deserts-and-xeric-shrublands on 21 May 2019
Acknowledgements
We are grateful with L. Eguiarte-Fruns and R. López Wilchis for discussions throughout the entire project, with R. Medina and Ó. Romero-Báez for assistance with genomic analyses, and with J. Searle and the people from his laboratory for their help and friendship during GP-S scientific visit. We deeply thank the Instituto de Ecología A.C. and Mr. Francisco Herrera and Mrs. Ernestina Rojas for their devoted support at the Mapimí Field Station. Our gratitude with A. Flores-Manzanero, T. Garrido-Garduño, M. Suárez-Atilano and M. Luna-Bárcenas for their enthusiastic help during fieldwork, S. Castañeda‐Rico and T. Garrido‐Garduño for molecular advice, and A. González and M. Baltazar for computational support. We truly thank the anonymous reviewers which helped us improve the manuscript. GP-S had a scholarship provided by CONACyT (becario 661803) for her Master’s in the Programa de Maestría en Ciencias Biológicas de la Universidad Nacional Autónoma de México (UNAM) and financial support from Programa de Estudios de Posgrado (PAEP 2017 and 2018). The project had financial support from Programa de Apoyo a Proyectos de Investigación e Innovación Tenconlógica granted to EV-D (research grant Papiit IN201716). EV-D received financial support for a sabbatical at the Estación Biológica de Doñana-CSIC from Dirección General de Asuntos del Personal Académico (DGAPA, UNAM; PASPA No. 067/2023) and Consejo Nacional de Humanidades, Ciencias y Tecnologias (CONAHCyT).
Author information
Authors and Affiliations
Contributions
EV-D conceived the original idea, designed the research and obtained the financial support. EV-D and GP-S conducted the fieldwork. GP-S performed the laboratory work. EV-D and GP-S performed the analyses. EV-D wrote the manuscript and GP-S agreed on the final version and submission of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Research ethics statement
Procedures were conducted in strict accordance with the guidelines of the American Society of Mammalogists for use of wild mammal species (Sikes 2016) and with the corresponding collecting permits (FAUT 0168). No institutional ethical approval was required given no experimental procedures or killing of individuals were performed.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Giorgio Bertorelle.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pineda-Sánchez, G., Vázquez-Domínguez, E. Desert landscape features influencing the microgeographic genetic structure of Nelson’s pocket mouse Chaetodipus nelsoni. Heredity 134, 21–32 (2025). https://doi.org/10.1038/s41437-024-00732-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41437-024-00732-y