Abstract
To explore the effect of aging, a cohort study is being performed on Sado Island, which is located in the Sea of Japan. Sado Island is close to the eastern coast of Japan, yet its population speaks the western Japanese dialect. Consequently, the genetic background of the population of Sado Island is of interest. Based on Nei’s genetic distance, we compared the allele frequencies of people from Sado Island to those of people from Nagahama and Miyagi, which are located in the western and northeastern parts of Honshu, respectively. The results showed that the populations of Miyagi and Nagahama are genetically closer to each other than to the population of Sado Island. Because the Sado and Honshu Islands are isolated by a channel, it is possible that genetic drift occurred within Sado Island, which would explain the uniqueness of the people of this region.
Similar content being viewed by others
Japan is experiencing a “super-aging” society1. To explore the effect of aging, a cross-sectional study was performed, which was a subanalysis of the Project in Sado for Total Health (PROST), a hospital-based cohort study in Sado City, Niigata Prefecture, Japan (http://square.umin.ac.jp/prost/). PROST, a cohort study targeting outpatients (age ≥20 years) of Sado General Hospital2, began in June 2008 and is currently ongoing.
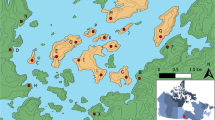
Sado Island is located 30 km off the coast of Niigata City, which is on the eastern coast of Honshu, mainland Japan. Despite their proximity, the dialect spoken on Sado Island is different from that spoken in Niigata3. Figure 1 shows the location of Sado Island and the distribution of Japanese dialects summarized in a previous study4. We hypothesized that the cultural difference reflects a difference in the ancestors of these populations. In this study, we investigated the genetic distances between the population of Sado Island and populations in eastern and western Japan.
This project was part of the PROST study, and it received the approval of the ethics committee of Niigata University School of Medicine (No. G2015-0811) as well as the ethics committee of Tohoku Medical Megabank Project (No. 2018-4-78). The samples used were obtained from the cohort participants, who gave their written consent. In addition, genotype data were securely controlled under the Materials and Information Distribution Review Committee of Tohoku Medical Megabank Project, and data sharing with researchers was discussed with the review committee. (https://www.megabank.tohoku.ac.jp/wp/wp-content/uploads/2018/05/2017-1014.pdf).
We used the Japonica SNP array5 to genotype the participants in PROST according to the manufacturer’s instructions. In the present study, 1750 individuals were genotyped. All genotyped samples passed the recommended sample quality control metric for the AXIOM arrays (dish QC40.82); we excluded three control samples with an overall call rate of <99%. We recalled the remaining 1747 samples with Genotyping Console 4.2.0.26 software (Affymetrix). We performed LD pruning using PLINK 2.00 alpha with the option “–indep-pairwise 50 10 0.1.” We also excluded loci when their minor allele frequencies (MAF) were lower than 5% and when the loci failed (P < 0.05) the Hardy–Weinberg test. In total, 5,078 loci passed these filters.
As a representative of the western Japan population, allele frequencies were obtained from the Human Genetic Variation Database6, which contains the genotype counts of participants in The Nagahama Prospective Genome Cohort for the Comprehensive Human Bioscience7. In the present study, this dataset is referred to as Nagahama. As a representative of the eastern Japan population, we used iJGVD8, which contains data obtained from the Miyagi prefecture9. This dataset is referred to as Miyagi in this study. The locations of Miyagi and Nagahama are shown in Fig. 1. Note that both Miyagi and Nagahama are located in Honshu.
Nei’s genetic distance10 was calculated among the populations using all loci, and 99% confidence intervals of the Dst were calculated by bootstrap resampling, with 5,000 replications.
Table 1 shows the genetic distances and their confidence intervals (99%) among the populations of Sado, Miyagi, and Nagahama. The results showed that the genetic distances between the population on Sado Island and the Honshu populations were significantly larger than that between the Miyagi and Nagahama populations.
In addition, Table 1 shows that the genetic distance between the Sado and Nagahama populations was not significantly different from that between the Sado and Miyagi populations. Thus, the hypothesis that the Sado population is genetically closer to the population in western Japan than the population in eastern Japan was neither rejected nor supported. We confirmed that the participants in PROST on Sado Island were genetically closer to Japanese living in Tokyo (JPT) than to Han Chinese in Beijing (CHB) and Southern Chinese (CHS) using principal component analysis (PCA) (Supplementary Fig. 1).
To estimate the degree of isolation of the Sado population, we estimated the inbreeding coefficients (IBC) based on the SNP homozygosity of samples using plink version 1.90b6.9. The same set of SNPs originally selected for PCA was used (Supplementary Fig. 1). The means ± SDs of the estimated IBC of the Sado and JPT populations are −0.0041 ± 0.0656 and −0.0043 ± 0.0129, respectively. The difference in IBC between the two populations was not statistically significant (t-test, P > 0.05). In other words, the size of the population on Sado Island is not overly small.
This study shows that geographic distance and linguistic similarity do not reflect the genetic differences among these populations. In addition, genetic drift may be more substantial than the linguistic shift.
References
Muramatsu, N. & Akiyama, H. Japan: super-aging society preparing for the future. Gerontologist 51, 425–432 (2011).
Kitamura, K. et al. Modifiable factors associated with cognitive impairment in 1,143 Japanese Outpatients: the Project in Sado for Total Health (PROST). Dement. Geriatr. Cogn. Dis. Extra. 6, 341–349 (2016).
Fukushima, C. in The future of dialects: Selected papers from Methods in Dialectology XV (eds Marie-Hélène Côté, Remco Knooihuizen & John Nerbonne) (Language Science Press, 2016).
Onishi, T. Dialect research at the National Institute for Japanese Language. Dialectologia et Geolinguistica, 87–96 (2002).
Kawai, Y. et al. Japonica array: improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J. Hum. Genet. 60, 581–587 (2015).
Higasa, K. et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. J. Hum. Genet. 61, 547–553 (2016).
Tabara, Y. et al. Association of serum-free fatty acid level with reduced reflection pressure wave magnitude and central blood pressure: the Nagahama study. Hypertension 64, 1212–1218 (2014).
Yamaguchi-Kabata, Y. et al. iJGVD: an integrative Japanese genome variation database based on whole-genome sequencing. Hum. Genome. Var. 2, 15050 (2015).
Nagasaki, M. et al. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat. Commun. 6, 8018 (2015).
Nei, M. Genetic distance between populations. Am. Nat. 106, 283–292 (1972).
Acknowledgements
We thank Professor Yoichi Furukawa and an anonymous reviewer for their useful comments and suggestions. This work was supported by JSPS Kakenhi (JP17K08682).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Misawa, K., Watanabe, H., Yokoseki, A. et al. Is the population of Sado Island genetically close to the population of western Japan?. Hum Genome Var 6, 26 (2019). https://doi.org/10.1038/s41439-019-0058-6
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41439-019-0058-6