Fig. 1: Flowchart of CaSpER algorithm.
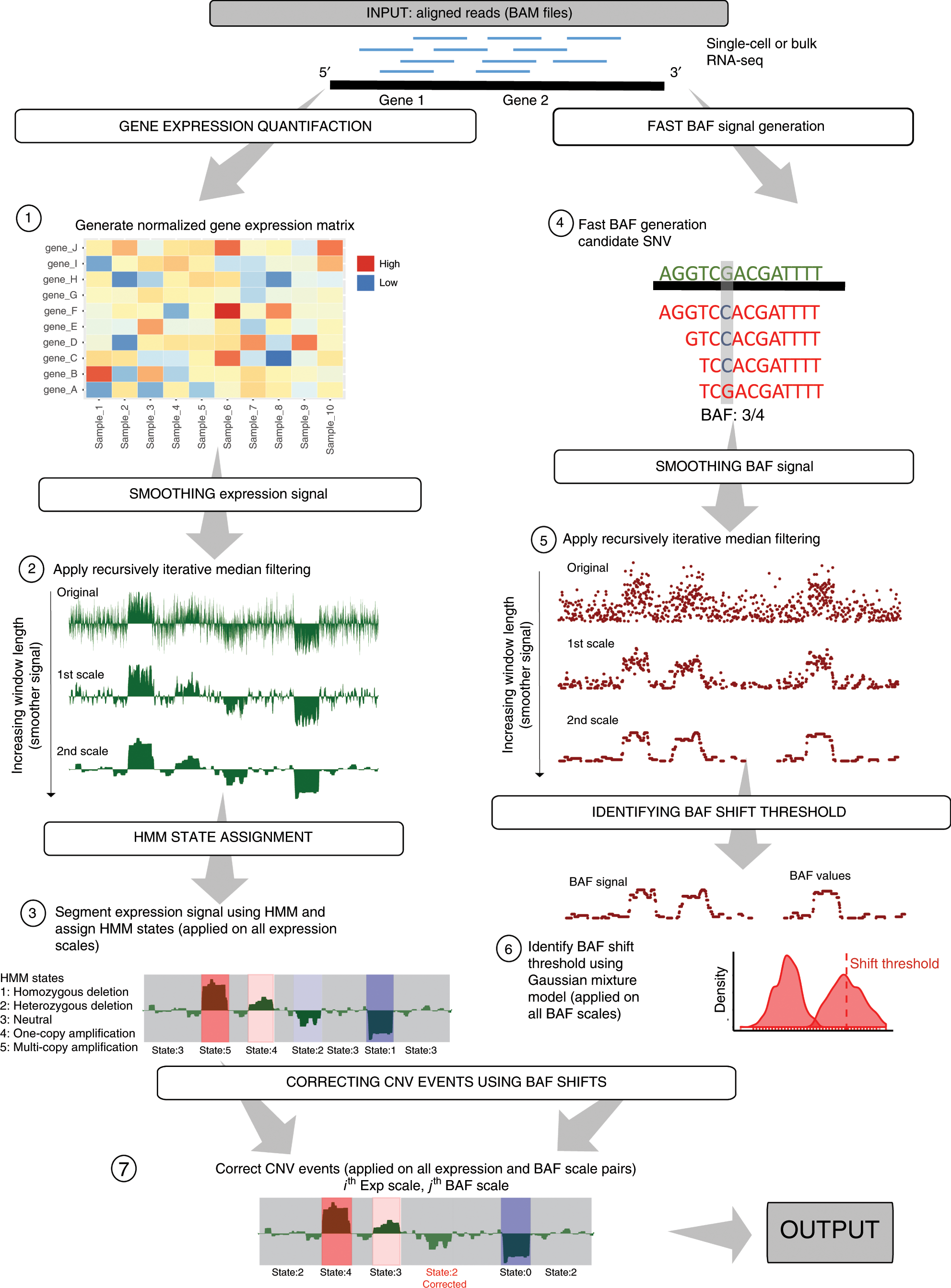
The CaSpER algorithm uses expression values and B-allele frequencies (BAF) from RNA-seq reads to estimate CNV events. A normalized gene expression matrix is generated (Step 1). Expression signal is smoothed by applying recursive iterative median filtering. Three-scale resolution of the expression signal is computed. (Step 2). For the smoothed signal at each scale, HMM is used to assign CNV states to regions and segment the signal into regions of similar copy number states (Step 3). Five CNV states are used in HMM model; 1: homozygous deletion, 2: heterozygous deletion, 3: neutral, 4: one-copy amplification, 5: multi-copy amplification. BAF information incorporated into the segmented CNV events. BAF information is extracted from mapped RNA-seq reads using an optimized BAF generation algorithm (Step 4). BAF signal is smoothed by applying recursive iterative median filtering. Three-scale resolution of the allele-based frequency signal is computed (Step 5). BAF shift threshold is estimated using a Gaussian mixture (Step 6). CNV events are corrected using BAF shifts and final CNV correction is applied to all the CNV and BAF scale pair combinations (Step 7).
