Fig. 1: Workflow of ribosome profiling.
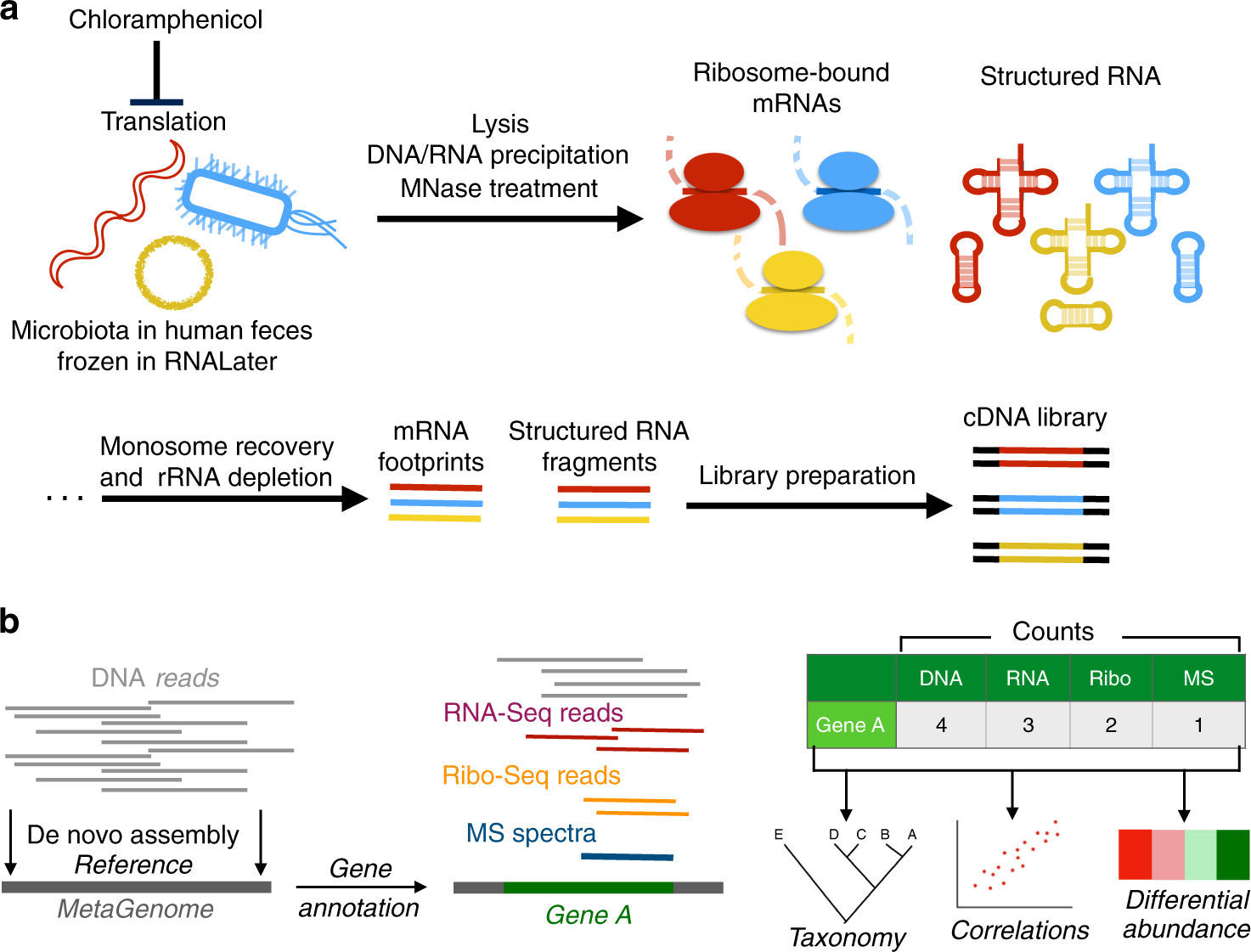
a Experimental workflow of MetaRibo-Seq. Chloramphenicol halts translation, bacteria within the community are lysed, MNase is used to create footprints, and footprints are converted to sequencing libraries. b Computational workflow of the multi-omics approach. De novo assemblies are created and annotated, predicted genes are quantified at multi-omic levels, and taxonomy, correlations, and differential abundance are determined from these results.
