Fig. 1: Overview for using aptardi.
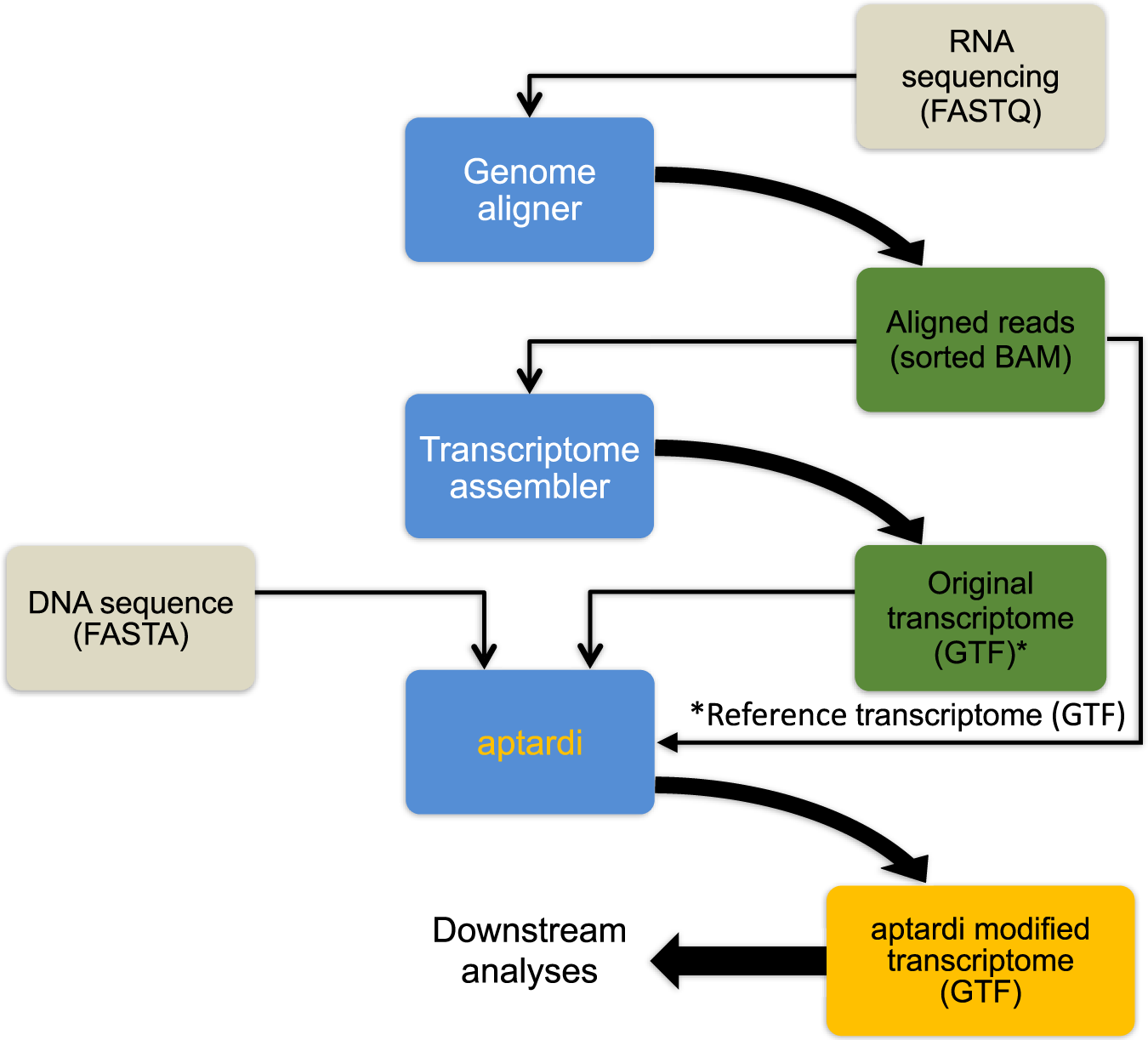
Aptardi requires three files as input: (1) FASTA file of DNA sequence with headers by chromosome, (2) sorted Binary Alignment Map (BAM) file of reads aligned to the genome, and (3) General Feature Format (GTF) file of transcript structures. Blue boxes represent software. Yellow writing/boxes indicate aptardi incorporation. Note transcript structures can be derived from a reference transcriptome (i.e., Ensembl annotation) in lieu of the original transcriptome generated from a transcriptome assembler.
