Abstract
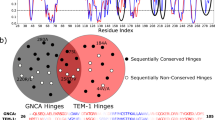
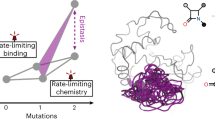
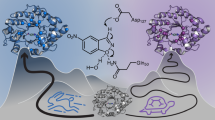
Our ability to understand protein evolution hinges on understanding how evolutionary landscapes are shaped at the fundamental protein level. Using TEM-1 β-lactamase we show that molecular traits related to the statistical ensemble nature of protein structure contribute to broader substrate specificity, active site-scaffold communication, and the selection of stabilizing substitutions. During the evolution of cefotaxime resistance, the initial mutation reorganizes the active site, introducing a new function conformation. Secondary substitutions improve catalytic efficiency by redistributing the ensemble and restoring a significant population of the original conformation, rather than by stabilizing the new conformation. Stability defects associated with initial mutations are not evenly disseminated but are clustered at specific distal scaffold elements. The capacity of mutants to independently modulate the populations of individual active site walls and scaffold regions through narrow residue networks, produces conformational epistasis and a combinatorial set of cefotaximase states, which enables local compensation of scaffold defects.
Similar content being viewed by others
Data availability
The HDX-MS data generated in this study have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository under accession code PXD069558. Previously published BMRB codes utilized: BMRB 7236. Source data are provided in the source data file. Materials used are available from the corresponding author.
References
Salverda, M. L., De Visser, J. A. & Barlow, M. Natural evolution of TEM-1 beta-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol. Rev. 34, 1015–1036 (2010).
Goldsmith, M. & Tawfik, D. S. Enzyme engineering: reaching the maximal catalytic efficiency peak. Curr. Opin. Struct. Biol. 47, 140–150 (2017).
Kaltenbach, M. & Tokuriki, N. Dynamics and constraints of enzyme evolution. J. Exp. Zool. B Mol. Dev. Evol. 322, 468–487 (2014).
Miton, C. M. & Tokuriki, N. How mutational epistasis impairs predictability in protein evolution and design. Protein Sci. 25, 1260–1272 (2016).
Tokuriki, N., Stricher, F., Serrano, L. & Tawfik, D. S. How protein stability and new functions trade off. PLoS Comput Biol. 4, e1000002 (2008).
Wang, X., Minasov, G. & Shoichet, B. K. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 320, 85–95 (2002).
Toth-Petroczy, A. & Tawfik, D. S. The robustness and innovability of protein folds. Curr. Opin. Struct. Biol. 26, 131–138 (2014).
Aharoni, A. et al. The evolvability of promiscuous protein functions. Nat. Genet 37, 73–76 (2005).
Johansson, K. E. & Lindorff-Larsen, K. Structural heterogeneity and dynamics in protein evolution and design. Curr. Opin. Struct. Biol. 48, 157–163 (2018).
Khersonsky, O., Roodveldt, C. & Tawfik, D. S. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10, 498–508 (2006).
Matsumura, I. & Ellington, A. D. In vitro evolution of beta-glucuronidase into a beta-galactosidase proceeds through non-specific intermediates. J. Mol. Biol. 305, 331–339 (2001).
Pabis, A., Risso, V. A., Sanchez-Ruiz, J. M. & Kamerlin, S. C. Cooperativity and flexibility in enzyme evolution. Curr. Opin. Struct. Biol. 48, 83–92 (2018).
Petrovic, D., Risso, V. A., Kamerlin, S. C. L. & Sanchez-Ruiz, J. M. Conformational dynamics and enzyme evolution. J. R. Soc. Interface. https://doi.org/10.1098/rsif.2018.0330 (2018).
Tokuriki, N. & Tawfik, D. S. Protein dynamism and evolvability. Science 324, 203–207 (2009).
Wouters, M. A., Liu, K., Riek, P. & Husain, A. A despecialization step underlying evolution of a family of serine proteases. Mol. Cell 12, 343–354 (2003).
Bloom, J. D., Labthavikul, S. T., Otey, C. R. & Arnold, F. H. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA 103, 5869–5874 (2006).
Guerois, R., Nielsen, J. E. & Serrano, L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 320, 369–387 (2002).
Schymkowitz, J. et al. The FoldX web server: an online force field. Nucleic Acids Res 33, W382–W388 (2005).
Tokuriki, N., Stricher, F., Schymkowitz, J., Serrano, L. & Tawfik, D. S. The stability effects of protein mutations appear to be universally distributed. J. Mol. Biol. 369, 1318–1332 (2007).
Axe, D. D., Foster, N. W. & Fersht, A. R. A search for single substitutions that eliminate enzymatic function in a bacterial ribonuclease. Biochemistry 37, 7157–7166 (1998).
Drummond, D. A., Silberg, J. J., Meyer, M. M., Wilke, C. O. & Arnold, F. H. On the conservative nature of intragenic recombination. Proc. Natl. Acad. Sci. USA 102, 5380–5385 (2005).
Guo, H. H., Choe, J. & Loeb, L. A. Protein tolerance to random amino acid change. Proc. Natl. Acad. Sci. USA 101, 9205–9210 (2004).
Rennell, D., Bouvier, S. E., Hardy, L. W. & Poteete, A. R. Systematic mutation of bacteriophage T4 lysozyme. J. Mol. Biol. 222, 67–88 (1991).
Shafikhani, S., Siegel, R. A., Ferrari, E. & Schellenberger, V. Generation of large libraries of random mutants in Bacillus subtilis by PCR-based plasmid multimerization. Biotechniques 23, 304–310 (1997).
Tokuriki, N. & Tawfik, D. S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 19, 596–604 (2009).
Bershtein, S., Segal, M., Bekerman, R., Tokuriki, N. & Tawfik, D. S. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 444, 929–932 (2006).
Bloom, J. D., Arnold, F. H. & Wilke, C. O. Breaking proteins with mutations: threads and thresholds in evolution. Mol. Syst. Biol. 3, 76 (2007).
Bloom, J. D. et al. Thermodynamic prediction of protein neutrality. Proc. Natl. Acad. Sci. USA 102, 606–611 (2005).
Dellus-Gur, E., Toth-Petroczy, A., Elias, M. & Tawfik, D. S. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J. Mol. Biol. 425, 2609–2621 (2013).
Bershtein, S., Goldin, K. & Tawfik, D. S. Intense neutral drifts yield robust and evolvable consensus proteins. J. Mol. Biol. 379, 1029–1044 (2008).
Gaucher, E. A., Govindarajan, S. & Ganesh, O. K. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451, 704–707 (2008).
Godoy-Ruiz, R., Perez-Jimenez, R., Ibarra-Molero, B. & Sanchez-Ruiz, J. M. A stability pattern of protein hydrophobic mutations that reflects evolutionary structural optimization. Biophys. J. 89, 3320–3331 (2005).
Bornberg-Bauer, E. & Chan, H. S. Modeling evolutionary landscapes: mutational stability, topology, and superfunnels in sequence space. Proc. Natl. Acad. Sci. USA 96, 10689–10694 (1999).
Starr, T. N. & Thornton, J. W. Epistasis in protein evolution. Protein Sci. 25, 1204–1218 (2016).
Knies, J. L., Cai, F. & Weinreich, D. M. Enzyme efficiency but not thermostability drives cefotaxime resistance evolution in TEM-1 beta-Lactamase. Mol. Biol. Evol. 34, 1040–1054 (2017).
Kather, I., Jakob, R. P., Dobbek, H. & Schmid, F. X. Increased folding stability of TEM-1 beta-lactamase by in vitro selection. J. Mol. Biol. 383, 238–251 (2008).
Lim, S. A., Hart, K. M., Harms, M. J. & Marqusee, S. Evolutionary trend toward kinetic stability in the folding trajectory of RNases H. Proc. Natl. Acad. Sci. USA 113, 13045–13050 (2016).
Romero-Romero, M. L. et al. Selection for protein kinetic stability connects denaturation temperatures to organismal temperatures and provides clues to archaean life. PLoS One 11, e0156657 (2016).
Yu, H. & Dalby, P. A. Exploiting correlated molecular-dynamics networks to counteract enzyme activity-stability trade-off. Proc. Natl. Acad. Sci. USA 115, E12192–E12200 (2018).
Yu, H. & Dalby, P. A. Coupled molecular dynamics mediate long- and short-range epistasis between mutations that affect stability and aggregation kinetics. Proc. Natl. Acad. Sci. USA 115, E11043–E11052 (2018).
Ose, N. J., Campitelli, P., Patel, R., Kumar, S. & Ozkan, S. B. Protein dynamics provide mechanistic insights about epistasis among common missense polymorphisms. Biophys. J. 122, 2938–2947 (2023).
Ambler, R. P. et al. A standard numbering scheme for the class A beta-lactamases. Biochem J. 276, 269–270 (1991).
Salverda, M. L. et al. Initial mutations direct alternative pathways of protein evolution. PLoS Genet 7, e1001321 (2011).
Palzkill, T. Structural and mechanistic basis for extended-spectrum drug-resistance mutations in altering the specificity of TEM, CTX-M, and KPC beta-lactamases. Front Mol. Biosci. 5, 16 (2018).
Orencia, M. C., Yoon, J. S., Ness, J. E., Stemmer, W. P. & Stevens, R. C. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat. Struct. Biol. 8, 238–242 (2001).
Dellus-Gur, E. et al. Negative epistasis and evolvability in TEM-1 beta-lactamase-the thin line between an enzyme’s conformational freedom and disorder. J. Mol. Biol. 427, 2396–2409 (2015).
Docquier, J. D., Benvenuti, M., Calderone, V., Rossolini, G. M. & Mangani, S. Structure of the extended-spectrum beta-lactamase TEM-72 inhibited by citrate. Acta Crystallogr Sect. F. Struct. Biol. Cryst. Commun. 67, 303–306 (2011).
Shimamura, T. et al. Acyl-intermediate structures of the extended-spectrum class A beta-lactamase, Toho-1, in complex with cefotaxime, cephalothin, and benzylpenicillin. J. Biol. Chem. 277, 46601–46608 (2002).
Doucet, N., Savard, P. Y., Pelletier, J. N. & Gagne, S. M. NMR investigation of Tyr105 mutants in TEM-1 beta-lactamase: dynamics are correlated with function. J. Biol. Chem. 282, 21448–21459 (2007).
Savard, P. Y. & Gagne, S. M. Backbone dynamics of TEM-1 determined by NMR: evidence for a highly ordered protein. Biochemistry 45, 11414–11424 (2006).
Clouthier, C. M. et al. Chimeric beta-lactamases: global conservation of parental function and fast time-scale dynamics with increased slow motions. PLoS One 7, e52283 (2012).
Morin, S. & Gagne, S. M. NMR dynamics of PSE-4 beta-lactamase: an interplay of ps-ns order and mus-ms motions in the active site. Biophys. J. 96, 4681–4691 (2009).
Sakhrani, V. V. et al. Toho-1 beta-lactamase: backbone chemical shift assignments and changes in dynamics upon binding with avibactam. J. Biomol. NMR 75, 303–318 (2021).
Rossi, M. A., Palzkill, T., Almeida, F. C. L. & Vila, A. J. Slow Protein Dynamics Elicits New Enzymatic Functions by Means of Epistatic Interactions. Mol. Biol. Evol. https://doi.org/10.1093/molbev/msac194 (2022).
Elings, W. et al. Two beta-lactamase variants with reduced clavulanic acid inhibition display different millisecond dynamics. Antimicrob. Agents Chemother. 65, e0262820 (2021).
Huang, W., Petrosino, J., Hirsch, M., Shenkin, P. S. & Palzkill, T. Amino acid sequence determinants of beta-lactamase structure and activity. J. Mol. Biol. 258, 688–703 (1996).
Gobeil, S. M. C. et al. Maintenance of native-like protein dynamics may not be required for engineering functional proteins. Chem. Biol. 21, 1330–1340 (2014).
Gobeil, S. M. C. et al. The structural dynamics of engineered beta-lactamases vary broadly on three timescales yet sustain native function. Sci. Rep. 9, 6656 (2019).
Stemmer, W. P. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 (1994).
Hall, B. G. Predicting evolution by in vitro evolution requires determining evolutionary pathways. Antimicrob. Agents Chemother. 46, 3035–3038 (2002).
Weinreich, D. M., Delaney, N. F., Depristo, M. A. & Hartl, D. L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006).
Huang, W. & Palzkill, T. A natural polymorphism in beta-lactamase is a global suppressor. Proc. Natl. Acad. Sci. USA 94, 8801–8806 (1997).
Sideraki, V., Huang, W., Palzkill, T. & Gilbert, H. F. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc. Natl. Acad. Sci. USA 98, 283–288 (2001).
Steinberg, B. & Ostermeier, M. Environmental changes bridge evolutionary valleys. Sci. Adv. 2, e1500921 (2016).
Lee, S. et al. Natural and synthetic suppressor mutations defy stability-activity tradeoffs. Biochemistry 61, 398–407 (2022).
Horn, J. R. & Shoichet, B. K. Allosteric inhibition through core disruption. J. Mol. Biol. 336, 1283–1291 (2004).
Bowman, G. R., Bolin, E. R., Hart, K. M., Maguire, B. C. & Marqusee, S. Discovery of multiple hidden allosteric sites by combining Markov state models and experiments. Proc. Natl. Acad. Sci. USA 112, 2734–2739 (2015).
Bowman, G. R. & Geissler, P. L. Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites. Proc. Natl. Acad. Sci. USA 109, 11681–11686 (2012).
Meneksedag, D., Dogan, A., Kanlikilicer, P. & Ozkirimli, E. Communication between the active site and the allosteric site in class A beta-lactamases. Comput Biol. Chem. 43, 1–10 (2013).
Porter, J. R. et al. Cooperative changes in solvent exposure identify cryptic pockets, switches, and allosteric coupling. Biophys. J. 116, 818–830 (2019).
Beadle, B. M. & Shoichet, B. K. Structural bases of stability-function tradeoffs in enzymes. J. Mol. Biol. 321, 285–296 (2002).
Brown, K. M. et al. Compensatory mutations restore fitness during the evolution of dihydrofolate reductase. Mol. Biol. Evol. 27, 2682–2690 (2010).
Rodrigues, J. V. et al. Biophysical principles predict fitness landscapes of drug resistance. Proc. Natl. Acad. Sci. USA 113, E1470–E1478 (2016).
Chang, M. W. & Torbett, B. E. Accessory mutations maintain stability in drug-resistant HIV-1 protease. J. Mol. Biol. 410, 756–760 (2011).
Bloom, J. D., Gong, L. I. & Baltimore, D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328, 1272–1275 (2010).
Nezhad, N. G. et al. Recent advances in simultaneous thermostability-activity improvement of industrial enzymes through structure modification. Int J. Biol. Macromol. 232, 123440 (2023).
Teufl, M., Zajc, C. U. & Traxlmayr, M. W. Engineering strategies to overcome the stability-function trade-off in proteins. ACS Synth. Biol. 11, 1030–1039 (2022).
Blomberg, R. et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 503, 418–421 (2013).
Packer, M. S. & Liu, D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet 16, 379–394 (2015).
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007).
Frauenfelder, H., Sligar, S. G. & Wolynes, P. G. The energy landscapes and motions of proteins. Science 254, 1598–1603 (1991).
Luque, I., Leavitt, S. A. & Freire, E. The linkage between protein folding and functional cooperativity: two sides of the same coin? Annu Rev. Biophys. Biomol. Struct. 31, 235–256 (2002).
Hilser, V. J., Dowdy, D., Oas, T. G. & Freire, E. The structural distribution of cooperative interactions in proteins: analysis of the native state ensemble. Proc. Natl. Acad. Sci. USA 95, 9903–9908 (1998).
Hilser, V. J. & Freire, E. Structure-based calculation of the equilibrium folding pathway of proteins. Correlation with hydrogen exchange protection factors. J. Mol. Biol. 262, 756–772 (1996).
Motlagh, H. N., Wrabl, J. O., Li, J. & Hilser, V. J. The ensemble nature of allostery. Nature 508, 331–339 (2014).
Corbella, M., Pinto, G. P. & Kamerlin, S. C. L. Loop dynamics and the evolution of enzyme activity. Nat. Rev. Chem. 7, 536–547 (2023).
Campbell, E. et al. The role of protein dynamics in the evolution of new enzyme function. Nat. Chem. Biol. 12, 944–950 (2016).
Biel, J. T., Thompson, M. C., Cunningham, C. N., Corn, J. E. & Fraser, J. S. Flexibility and design: conformational heterogeneity along the evolutionary trajectory of a redesigned ubiquitin. Structure 25, 739–749 e733 (2017).
Campitelli, P., Modi, T., Kumar, S. & Ozkan, S. B. The role of conformational dynamics and allostery in modulating protein evolution. Annu Rev. Biophys. 49, 267–288 (2020).
Gonzalez, M. M., Abriata, L. A., Tomatis, P. E. & Vila, A. J. Optimization of conformational dynamics in an epistatic evolutionary trajectory. Mol. Biol. Evol. 33, 1768–1776 (2016).
Jimenez-Oses, G. et al. The role of distant mutations and allosteric regulation on LovD active site dynamics. Nat. Chem. Biol. 10, 431–436 (2014).
Risso, V. A. et al. De novo active sites for resurrected Precambrian enzymes. Nat. Commun. 8, 16113 (2017).
Saves, I. et al. Mass spectral kinetic study of acylation and deacylation during the hydrolysis of penicillins and cefotaxime by beta-lactamase TEM-1 and the G238S mutant. Biochemistry 34, 11660–11667 (1995).
Alejaldre, L. et al. Known evolutionary paths are accessible to engineered ss-lactamases having altered protein motions at the timescale of catalytic turnover. Front Mol. Biosci. 7, 599298 (2020).
Hart, K. M., Ho, C. M., Dutta, S., Gross, M. L. & Bowman, G. R. Modelling proteins’ hidden conformations to predict antibiotic resistance. Nat. Commun. 7, 12965, (2016).
Knoverek, C. R. et al. Opening of a cryptic pocket in beta-lactamase increases penicillinase activity. Proc Natl Acad Sci USA 118, https://doi.org/10.1073/pnas.2106473118 (2021).
Freire, E. The propagation of binding interactions to remote sites in proteins: analysis of the binding of the monoclonal antibody D1.3 to lysozyme. Proc. Natl. Acad. Sci. USA 96, 10118–10122 (1999).
Lockless, S. W. & Ranganathan, R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286, 295–299 (1999).
Liu, T., Whitten, S. T. & Hilser, V. J. Functional residues serve a dominant role in mediating the cooperativity of the protein ensemble. Proc. Natl. Acad. Sci. USA 104, 4347–4352 (2007).
Clarkson, M. W., Gilmore, S. A., Edgell, M. H. & Lee, A. L. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry 45, 7693–7699 (2006).
Hilser, V. J., Wrabl, J. O. & Motlagh, H. N. Structural and energetic basis of allostery. Annu Rev. Biophys. 41, 585–609 (2012).
Popovych, N., Sun, S., Ebright, R. H. & Kalodimos, C. G. Dynamically driven protein allostery. Nat. Struct. Mol. Biol. 13, 831–838 (2006).
Chen, Y., Delmas, J., Sirot, J., Shoichet, B. & Bonnet, R. Atomic resolution structures of CTX-M beta-lactamases: extended spectrum activities from increased mobility and decreased stability. J. Mol. Biol. 348, 349–362 (2005).
Hecky, J. & Muller, K. M. Structural perturbation and compensation by directed evolution at physiological temperature leads to thermostabilization of beta-lactamase. Biochemistry 44, 12640–12654 (2005).
Vanhove, M., Raquet, X. & Frere, J. M. Investigation of the folding pathway of the TEM-1 beta-lactamase. Proteins 22, 110–118 (1995).
Lejeune, A., Pain, R. H., Charlier, P., Frere, J. M. & Matagne, A. TEM-1 beta-lactamase folds in a nonhierarchical manner with transient non-native interactions involving the C-terminal region. Biochemistry 47, 1186–1193 (2008).
Schoene, C., Fierer, J. O., Bennett, S. P. & Howarth, M. SpyTag/SpyCatcher cyclization confers resilience to boiling on a mesophilic enzyme. Angew. Chem. Int Ed. Engl. 53, 6101–6104 (2014).
Iwai, H. & Pluckthun, A. Circular beta-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 459, 166–172 (1999).
Gonzalez, L. J., Bahr, G., Gonzalez, M. M., Bonomo, R. A. & Vila, A. J. In-cell kinetic stability is an essential trait in metallo-beta-lactamase evolution. Nat. Chem. Biol. 19, 1116–1126 (2023).
Liu, Q., Xun, G. & Feng, Y. The state-of-the-art strategies of protein engineering for enzyme stabilization. Biotechnol. Adv. 37, 530–537 (2019).
Long, D., Liu, M. & Yang, D. Accurately probing slow motions on millisecond timescales with a robust NMR relaxation experiment. J. Am. Chem. Soc. 130, 2432–2433 (2008).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Lipari, G. & Szabo, A. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules .1. theory and range of validity. J. Am. Chem. Soc. 104, 4546–4559 (1982).
d’Auvergne, E. J. & Gooley, P. R. Optimisation of NMR dynamic models II. a new methodology for the dual optimisation of the model-free parameters and the Brownian rotational diffusion tensor. J. Biomol. NMR 40, 121–133 (2008).
d’Auvergne, E. J. & Gooley, P. R. Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J. Biomol. NMR 40, 107–119 (2008).
Akaike, H. New look at statistical-model identification. IEEE T Autom. Contr Ac19, 716–723 (1974).
d’Auvergne, E. J. & Gooley, P. R. The use of model selection in the model-free analysis of protein dynamics. J. Biomol. NMR 25, 25–39 (2003).
Luz, Z. & Meiboom, S. Nuclear Magnetic Resonance Study of Protolysis of Trimethylammonium Ion in Aqueous Solution - Order of Reaction with Respect to Solvent. J. Chem. Phys. 39, 366-& (1963).
Carver, J. P. & Richards, R. E. General 2-site solution for chemical exchange produced dependence of T2 upon Carr-Purcell pulse separation. J. Magn. Reson 6, 89-& (1972).
Acknowledgements
We are grateful to Dr. Youlin Xia (St. Jude Children’s Reseach Hospital) for providing the CPMG pulse sequences, Dr. Todd Rappe (Minnesota NMR Center) for assistance with the relaxation experiments and Sofia Gonzalez for assistance with sample preparations. NMR experiments were carried out at USF’s Florida Center of Excellence for Drug Discovery and Innovation and the Minnesota NMR Center. This work was supported by the US National Institutes of Health (GM115854 to I.G. and AI161762 to Y.C.).
Author information
Authors and Affiliations
Contributions
E.A., D.K., Y.C. and I.G. designed research; E.A., D.K. and M.P. prepared samples; E.A., D.K., V.K. and J.M.D. acquired and analyzed NMR data; E.A., R.A. and I.G. analyzed HDX-MS data; L.J. performed enzymatic assays; S.M., Y.C. and I.G. provided resources; D.K. and I.G. drafted the manuscript; E.A., D.K., M.P., J.M.D., Y.C. and I.G. reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Arcia, E., Keramisanou, D., Jacobs, L.M.C. et al. Dynamic signature of activity-stability tradeoff in lactamase evolution. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68620-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68620-z