Abstract
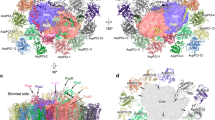
Prochlorococcus is a major contributor to primary production, and globally the most abundant photosynthetic genus of picocyanobacteria because it can adapt to highly stratified low-nutrient conditions that are characteristic of the surface ocean. Here, we examine the structural adaptations of the photosynthetic thylakoid membrane that enable different Prochlorococcus ecotypes to occupy high-light, low-light and nutrient-poor ecological niches. We used atomic force microscopy to image the different photosystem I (PSI) membrane architectures of the MED4 (high-light) Prochlorococcus ecotype grown under high-light and low-light conditions in addition to the MIT9313 (low-light) and SS120 (low-light) Prochlorococcus ecotypes grown under low-light conditions. Mass spectrometry quantified the relative abundance of PSI, photosystem II (PSII) and cytochrome b6f complexes and the various Pcb proteins in the thylakoid membrane. Atomic force microscopy topographs and structural modelling revealed a series of specialized PSI configurations, each adapted to the environmental niche occupied by a particular ecotype. MED4 PSI domains were loosely packed in the thylakoid membrane, whereas PSI in the low-light MIT9313 is organized into a tightly packed pseudo-hexagonal lattice that maximizes harvesting and trapping of light. There are approximately equal levels of PSI and PSII in MED4 and MIT9313, but nearly twofold more PSII than PSI in SS120, which also has a lower content of cytochrome b6f complexes. SS120 has a different tactic to cope with low-light levels, and SS120 thylakoids contained hundreds of closely packed Pcb–PSI supercomplexes that economize on the extra iron and nitrogen required to assemble PSI-only domains. Thus, the abundance and widespread distribution of Prochlorococcus reflect the strategies that various ecotypes employ for adapting to limitations in light and nutrient levels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://proteomecentral.proteomexchange.org) with the data set identifier PXD013506. All other data can be obtained from the corresponding author upon request. The following figures have associated raw data: Fig. 7, Supplementary Figs. 2–5.
References
Flombaum, P. et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl Acad. Sci. USA 110, 9824–9829 (2013).
Huston, M. A. & Wolverton, S. The global distribution of net primary production: resolving the paradox. Ecol. Monogr. 79, 343–377 (2009).
Partensky, F. & Garczarek, L. Prochlorococcus: advantages and limits of minimalism. Annu. Rev. Mar. Sci. 2, 305–331 (2010).
Goericke, R. & Repeta, D. J. The pigments of Prochlorococcus marinus: the presence of divinyl chlorophyll a and b in a marine procaryote. Limnol. Oceanogr. 37, 425–433 (1992).
Partensky, F., Hess, W. R. & Vaulot, D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106–127 (1999).
Partensky, F., Hoepffner, N., Li, W. & Ulloa, O. Photoacclimation of Prochlorococcus sp. (Prochlorophyta) strains isolated from the North Atlantic and the Mediterranean Sea. Plant Physiol. 101, 285–296 (1993).
Moore, L. R., Goericke, R. & Chisholm, S. W. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Ecol. Prog. Ser. 116, 259–275 (1995).
Moore, L. R., Goericke, R. & Chisholm, S. W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467 (1998).
Ferris, M. J. & Palenik, B. Niche adaptation in ocean cyanobacteria. Nature 396, 226–228 (1998).
Urbach, E., Scanlan, D. J., Distel, D. L., Waterbury, J. B. & Chisholm, S. W. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 46, 188–201 (1998).
West, N. J. & Scanlan, D. J. Niche-partitioning of Prochlorococcus populations in a stratified water column in the Eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65, 2585–2591 (1999).
Rocap, G., Distel, D. L., Waterbury, J. B. & Chisholm, S. W. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68, 1180–1191 (2002).
Zwirglmaier, K. et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161 (2008).
Martiny, A. C., Tai, A. P. K., Veneziano, D., Primeau, F. & Chisholm, S. W. Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ. Microbiol. 11, 823–832 (2009).
Malmstrom, R. R. et al. Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J. 4, 1252 (2010).
Biller, S. J., Berube, P. M., Lindell, D. & Chisholm, S. W. Prochlorococcus: the structure and function of collective diversity. Nat. Rev. Microbiol. 13, 13 (2015).
Moore, L. R. & Chisholm, S. W. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol. Oceanogr. 44, 628–638 (1999).
J. T. O. Kirk in Light and Photosynthesis in Aquatic Ecosystems Ch. 6 (Cambridge University Press, 1994).
Palenik, B. P. & Haselkorn, R. Multiple evolutionary origins of prochlorophytes, the chlorophyll b-containing prokaryotes. Nature 355, 265–267 (1992).
Urbach, E., Robertson, D. L. & Chisholm, S. W. Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature 355, 267 (1992).
Scanlan, D. J. & West, N. J. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40, 1–12 (2002).
Ting, C. S., Rocap, G., King, J. & Chisholm, S. W. Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10, 134–142 (2002).
La Roche, J. et al. Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc. Natl Acad. Sci. USA 93, 15244–15248 (1996).
Chen, M. & Bibby, T. S. Photosynthetic apparatus of antenna-reaction centres supercomplexes in oxyphotobacteria: insight through significance of Pcb/IsiA proteins. Photosynth. Res. 86, 165–173 (2005).
Bibby, T. S., Nield, J., Partensky, F. & Barber, J. Oxyphotobacteria: antenna ring around photosystem I. Nature 413, 590 (2001a).
Bibby, T. S., Mary, I., Nield, J., Partensky, F. & Barber, J. Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424, 1051 (2003).
Bibby, T. S., Nield, J. & Barber, J. Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412, 743 (2001b).
Boekema, E. J. et al. A giant chlorophyll–protein complex induced by iron deficiency in cyanobacteria. Nature 412, 745 (2001).
Ting, C. S., Hsieh, C., Sundararaman, S., Mannella, C. & Marko, M. Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J. Bacteriol. 189, 4485–4493 (2007).
Kirchhoff, H., Lenhert, S., Büchel, C., Chi, L. & Nield, J. Probing the organization of photosystem II in photosynthetic membranes by atomic force microscopy. Biochemistry 47, 431–440 (2008).
Sznee, K. et al. Jumping mode atomic force microscopy on grana membranes from spinach. J. Biol. Chem. 286, 39164–39171 (2011).
Johnson, M. P., Vasilev, C., Olsen, J. D. & Hunter, C. N. Nanodomains of cytochrome b6f and photosystem II complexes in spinach grana thylakoid membranes. Plant Cell 26, 3051–3061 (2014).
Onoa, B. et al. Atomic force microscopy of photosystem II and its unit cell clustering quantitatively delineate the mesoscale variability in Arabidopsis thylakoids. PloS ONE 9, e101470 (2014).
Phuthong, W. et al. The use of contact mode atomic force microscopy in aqueous medium for structural analysis of spinach photosynthetic complexes. Plant Physiol. 169, 1318–1332 (2015).
Tietz, S. et al. Functional implications of photosystem II crystal formation in photosynthetic membranes. J. Biol. Chem. 290, 14091–14106 (2015).
Wood, W. H. et al. Dynamic thylakoid stacking regulates the balance between linear and cyclic photosynthetic electron transfer. Nat. Plants 4, 116 (2018).
Liu, L. N. & Scheuring, S. Investigation of photosynthetic membrane structure using atomic force microscopy. Trends Plant Sci. 18, 277–286 (2013).
Kumar, S. et al. Direct imaging of protein organisation in an intact bacterial organelle using high-resolution atomic force microscopy. ACS Nano 11, 126–133 (2017).
MacGregor-Chatwin, C. et al. Lateral segregation of photosystem I in cyanobacterial thylakoids. Plant Cell 29, 1119–1136 (2017).
Casella, S. et al. Dissecting the native architecture and dynamics of cyanobacterial photosynthetic machinery. Mol. Plant 10, 1434–1448 (2017).
Jordan, P. et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411, 909 (2001).
Malavath, T., Caspy, I., Netzer-El, S. Y., Klaiman, D. & Nelson, N. Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis. Biochim. Biophys. Acta Bioenerg. 1859, 645–654 (2018).
Kubota-Kawai, H. et al. X-ray structure of an asymmetrical trimeric ferredoxin–photosystem I complex. Nat. Plants 4, 218–224 (2018).
Umena, Y., Kawakami, K., Shen, J. R. & Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55 (2011).
Waldbauer, J. R., Rodrigue, S., Coleman, M. L. & Chisholm, S. W. Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PloS ONE 7, e43432 (2012).
Domínguez-Martín, M. A. et al. Quantitative proteomics shows extensive remodeling induced by nitrogen limitation in Prochlorococcus marinus SS120. mSystems 2, e00008–e00017 (2017).
Jeffrey, S. T. & Humphrey, G. F. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. 167, 191–194 (1975).
Biggins, J. & Bruce, D. Regulation of excitation energy transfer in organisms containing phycobilins. Photosynth. Res. 20, 1–34 (1989).
Andersson, B. & Anderson, J. M. Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim. Biophys. Acta Bioenerg. 593, 427–440 (1980).
Engel, B. D. et al. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4, e04889 (2015).
Rippka, R. et al. Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. strain PCC 9511, the first axenic chlorophyll a2/b2-containing cyanobacterium (Oxyphotobacteria). Int. J. Syst. Evolut. Microbiol. 50, 1833–1847 (2000).
Bonisteel, E. M. et al. Strain specific differences in rates of Photosystem II repair in picocyanobacteria correlate to differences in FtsH protein levels and isoform expression patterns. PloS ONE 13, e0209115 (2018).
Nečas, D. & Klapetek, P. Gwyddion: an open-source software for SPM data analysis. Open Phys. 10, 181–188 (2012).
Mathematica v.11.3 (Wolfram Research, Inc., 2018).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Sener, M. K. et al. Excitation migration in trimeric cyanobacterial photosystem I. J. Chem. Phys. 120, 11183–11195 (2004).
Hitchcock, A. et al. Biosynthesis of chlorophyll a in a purple bacterial phototroph and assembly into a plant chlorophyll–protein complex. ACS Synth. Biol. 5, 948–954 (2016).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Fabre, B. et al. Comparison of label-free quantification methods for the determination of protein complexes subunits stoichiometry. EuPA Open Proteom. 4, 82–86 (2014).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Li, L. et al. Proteome dynamics reveals pro-inflammatory remodeling of plasma proteome in a mouse model of NAFLD. J. Proteome Res. 15, 3388–3404 (2016).
García-Plazaola, J. I. & Becerril, J. M. A rapid high-performance liquid chromatography method to measure lipophilic antioxidants in stressed plants: simultaneous determination of carotenoids and tocopherols. Phytochem. Anal. 10, 307–313 (1999).
Kalb, V. F. Jr & Bernlohr, R. W. A new spectrophotometric assay for protein in cell extracts. Anal. Biochem. 82, 362–371 (1977).
Acknowledgements
This work was supported by Advanced Award 338895 from the European Research Council which funded C.M.-C., P.J.J., J.W.C. and P.Q. and provided partial support for C.N.H. The authors C.N.H. and A.H. also gratefully acknowledge financial support from the Biotechnology and Biological Sciences Research Council (BBSRC UK), award number BB/M000265/1. C.N.H., M.S. and Z.L.-S. were supported by the Photosynthetic Antenna Research Center, an Energy Frontier Research Center funded by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under award number DE-SC 0001035. M.S. and Z.L.-S. were also supported by the National Science Foundation (grant no. MCB1616590) and the National Institutes of Health (grant no. 9P41GM104601). D.J.S. acknowledges funding from NERC (grant no. NE/N003241/1) and The Leverhulme Trust (grant no. RPG-2014-354). M.J.D. acknowledges support from the BBSRC UK (grant no. BB/M012166/1). M.P.J. would like to acknowledge BBSRC grant no. BB/P002005/1 and the Grantham Centres for Sustainable Futures, University of Sheffield, for G.E.M.’s studentship.
Author information
Authors and Affiliations
Contributions
C.M.-C., D.J.S. and C.N.H. designed the research. C.M.-C., P.J.J., M.S., J.W.C., A.H., P.Q., M.J.D., G.E.M. and D.J.S. performed the research. C.M.-C., M.S., M.P.J., Z.L.-S. and C.N.H. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature Plants thanks Douglas Campbell and other anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5, and Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
MacGregor-Chatwin, C., Jackson, P.J., Sener, M. et al. Membrane organization of photosystem I complexes in the most abundant phototroph on Earth. Nat. Plants 5, 879–889 (2019). https://doi.org/10.1038/s41477-019-0475-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-019-0475-z
This article is cited by
-
Hypothesis: bacteria live on the edge of phase transitions with a cell cycle regulated by a water-clock
Theory in Biosciences (2024)
-
Structural variability, coordination and adaptation of a native photosynthetic machinery
Nature Plants (2020)