Abstract
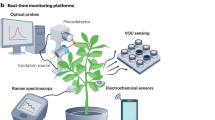
Innovative approaches are urgently required to alleviate the growing pressure on agriculture to meet the rising demand for food. A key challenge for plant biology is to bridge the notable knowledge gap between our detailed understanding of model plants grown under laboratory conditions and the agriculturally important crops cultivated in fields or production facilities. This Perspective highlights the recent development of new analytical tools that are rapid and non-destructive and provide tissue-, cell- or organelle-specific information on living plants in real time, with the potential to extend across multiple species in field applications. We evaluate the utility of engineered plant nanosensors and portable Raman spectroscopy to detect biotic and abiotic stresses, monitor plant hormonal signalling as well as characterize the soil, phytobiome and crop health in a non- or minimally invasive manner. We propose leveraging these tools to bridge the aforementioned fundamental gap with new synthesis and integration of expertise from plant biology, engineering and data science. Lastly, we assess the economic potential and discuss implementation strategies that will ensure the acceptance and successful integration of these modern tools in future farming practices in traditional as well as urban agriculture.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Godfray, H. C. J. et al. Food security: the challenge of feeding 9 billion people. Science 327, 812–818 (2010).
Gerten, D. et al. Feeding ten billion people is possible within four terrestrial planetary boundaries. Nat. Sustain. 3, 200–208 (2020).
King, T. et al. Food safety for food security: relationship between global megatrends and developments in food safety. Trends Food Sci. Tech. 68, 160–175 (2017).
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E. & Mittler, R. Abiotic and biotic stress combinations. New Phytol. 203, 32–43 (2014).
Gupta, A., Rico-Medina, A. & Caño-Delgado, A. I. The physiology of plant responses to drought. Science 368, 266–269 (2020).
Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019).
van Esse, H. P., Reuber, T. L. & van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 225, 70–86 (2020).
Lesk, C., Rowhani, P. & Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 529, 84–87 (2016).
Dai, A. Increasing drought under global warming in observations and models. Nat. Clim. Change 3, 52–58 (2013).
van Meijl, H. et al. Comparing impacts of climate change and mitigation on global agriculture by 2050. Environ. Res. Lett. 13, 064021 (2018).
Wiebe, K., Robinson, S. & Cattaneo, A. in Sustainable Food and Agriculture 55–74 (Elsevier, 2019).
Walter, A., Finger, R., Huber, R. & Buchmann, N. Smart farming is key to developing sustainable agriculture. Proc. Natl Acad. Sci. USA 114, 6148–6150 (2017).
Li, S. et al. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 560, 595–600 (2018).
Balmford, A. et al. The environmental costs and benefits of high-yield farming. Nat. Sustain. 1, 477–485 (2018).
Specht, K. et al. Urban agriculture of the future: an overview of sustainability aspects of food production in and on buildings. Agric. Human Values 31, 33–51 (2014).
Benke, K. & Tomkins, B. Future food-production systems: vertical farming and controlled-environment agriculture. Sustain. Sci. Pract. Policy 13, 13–26 (2017).
Torero Cullen, M. A battle plan for ensuring global food supplies during the COVID-19 crisis. FAO http://www.fao.org/news/story/en/item/1268059/icode/ (2020).
Berens, M. L. et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl Acad. Sci. USA 116, 2364–2373 (2019).
Riedlmeier, M. et al. Monoterpenes support systemic acquired resistance within and between plants. Plant Cell 29, 1440–1459 (2017).
Zhu, J. K. Abiotic stress signaling and responses in plants. Cell 167, 313–324 (2016).
Rhee, S. Y. et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31, 224–228 (2003).
Bevan, M. & Walsh, S. The Arabidopsis genome: a foundation for plant research. Genome Res. 15, 1632–1642 (2005).
Li, Z. & Sillanpää, M. J. Dynamic quantitative trait locus analysis of plant phenomic data. Trends Plant Sci. 20, 822–833 (2015).
Walia, A., Waadt, R. & Jones, A. M. Genetically encoded biosensors in plants: pathways to discovery. Annu. Rev. Plant Biol. 69, 497–524 (2018).
Borrill, P. Blurring the boundaries between cereal crops and model plants. New Phytol. 9, 16229 (2019).
Carpentler, S. C. et al. Proteome analysis of non-model plants: a challenging but powerful approach. Mass Spectrom. Rev. 27, 354–377 (2008).
Long, S. P., Marshall-Colon, A. & Zhu, X. G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66 (2015).
Jackson, S. A. Rice: the first crop genome. Rice 9, 1–3 (2016).
Yang, N. et al. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat. Genet. 51, 1052–1059 (2019).
Sant’Ana, D. V. P. & Lefsrud, M. Tomato proteomics: tomato as a model for crop proteomics. Scientia Horticulturae 239, 224–233 (2018).
Unamba, C. I. N., Nag, A. & Sharma, R. K. Next generation sequencing technologies: the doorway to the unexplored genomics of non-model plants. Front. Plant Sci. 6, 1074 (2015).
Richards, C. L. et al. Ecological plant epigenetics: evidence from model and non-model species, and the way forward. Ecol. Lett. 20, 1576–1590 (2017).
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L. & Fernie, A. R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1, 387–396 (2006).
Fernie, A. R., Trethewey, R. N., Krotzky, A. J. & Willmitzer, L. Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5, 763–769 (2004).
Lytovchenko, A. et al. Application of GC-MS for the detection of lipophilic compounds in diverse plant tissues. Plant Methods 5, 4 (2009).
Nagano, A. J. et al. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 151, 1358–1369 (2012).
Shakoor, N., Lee, S. & Mockler, T. C. High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 38, 184–192 (2017).
Araus, J. L. & Cairns, J. E. Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci. 19, 52–61 (2014).
Kirchgessner, N. et al. The ETH field phenotyping platform FIP: a cable-suspended multi-sensor system. Funct. Plant Biol. 44, 154 (2017).
Pieruschka, R. & Schurr, U. Plant phenotyping: past, present, and future. Plant Phenomics 2019, 7507131 (2019).
Yang, G. et al. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: current status and perspectives. Front. Plant Sci. 8, 1111 (2017).
Ampatzidis, Y. & Partel, V. UAV-based high throughput phenotyping in citrus utilizing multispectral imaging and artificial intelligence. Remote Sens. 11, 410 (2019).
Li, L., Zhang, Q. & Huang, D. A review of imaging techniques for plant phenotyping. Sensors 14, 20078–20111 (2014).
Chawade, A. et al. High-throughput field-phenotyping tools for plant breeding and precision agriculture. Agronomy 9, 258 (2019).
Zhao, C. et al. Crop phenomics: current status and perspectives. Front. Plant Sci. 10, 714 (2019).
Bai, G., Ge, Y., Hussain, W., Baenziger, P. S. & Graef, G. A multi-sensor system for high throughput field phenotyping in soybean and wheat breeding. Comput. Electron. Agric. 128, 181–192 (2016).
Cohen, Y., Alchanatis, V., Meron, M., Saranga, Y. & Tsipris, J. Estimation of leaf water potential by thermal imagery and spatial analysis. J. Exp. Bot. 56, 1843–1852 (2005).
Valle, B. et al. PYM: a new, affordable, image-based method using a Raspberry Pi to phenotype plant leaf area in a wide diversity of environments. Plant Methods 13, 1–17 (2017).
Gerhards, M., Schlerf, M., Mallick, K. & Udelhoven, T. Challenges and future perspectives of multi-/hyperspectral thermal infrared remote sensing for crop water-stress detection: a review. Remote Sens. 11, 1240 (2019).
Yang, W. et al. Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives. Mol. Plant 13, 187–214 (2020).
Fukatsu, T., Watanabe, T., Hu, H., Yoichi, H. & Hirafuji, M. Field monitoring support system for the occurrence of Leptocorisa chinensis Dallas (Hemiptera: Alydidae) using synthetic attractants, Field Servers, and image analysis. Comput. Electron. Agric. 80, 8–16 (2012).
Sadeghi-Tehran, P., Sabermanesh, K., Virlet, N. & Hawkesford, M. J. Automated method to determine two critical growth stages of wheat: heading and flowering. Front. Plant Sci. 8, 252 (2017).
Duan, L. et al. Novel digital features discriminate between drought resistant and drought sensitive rice under controlled and field conditions. Front. Plant Sci. 9, 492 (2018).
Jahnke, S. et al. phenoSeeder - a robot system for automated handling and phenotyping of individual seeds. Plant Physiol. 172, 1358–1370 (2016).
Mahlein, A. K. Plant disease detection by imaging sensors – parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 100, 241–254 (2016).
Alves Varella, C. A., Gleriani, J. M. & dos Santos, R. M. in Sugarcane: Agricultural Production, Bioenergy and Ethanol 185–203 (Elsevier Inc., 2015).
Tantalaki, N., Souravlas, S. & Roumeliotis, M. Data-driven decision making in precision agriculture: the rise of big data in agricultural systems. J. Agric. Food Inf. 20, 344–380 (2019).
Baumgart-Getz, A., Prokopy, L. S. & Floress, K. Why farmers adopt best management practice in the United States: a meta-analysis of the adoption literature. J. Environ. Manage. 96, 17–25 (2012).
Fujita, M. et al. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442 (2006).
Perilla-Henao, L. M. & Casteel, C. L. Vector-borne bacterial plant pathogens: interactions with hemipteran insects and plants. Front. Plant Sci. 7, 1163 (2016).
Bendix, C. & Lewis, J. D. The enemy within: phloem-limited pathogens. Mol. Plant Pathol. 19, 238–254 (2018).
Choi, W.-G. et al. Orchestrating rapid long-distance signaling in plants with Ca2+, ROS and electrical signals. Plant J. 90, 698–707 (2017).
Gilroy, S. et al. ROS, calcium, and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiol. 171, 1606–1615 (2016).
Shan, X., Yan, J. & Xie, D. Comparison of phytohormone signaling mechanisms. Curr. Opin. Plant Biol. 15, 84–91 (2012).
Zhao, Y., Qi, Z. & Berkowitz, G. A. Teaching an old hormone new tricks: cytosolic Ca2+ elevation involvement in plant brassinosteroid signal transduction cascades. Plant Physiol. 163, 555–565 (2013).
Seyfferth, C. & Tsuda, K. Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 5, 697 (2014).
Mohanta, T. K., Kumar, P. & Bae, H. Genomics and evolutionary aspect of calcium signaling event in calmodulin and calmodulin-like proteins in plants. BMC Plant Biol. 17, 38 (2017).
Huang, H., Ullah, F., Zhou, D. X., Yi, M. & Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 10, 800 (2019).
Wang, C., Liu, Y., Li, S. S. & Han, G. Z. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 167, 872–886 (2015).
Blázquez, M. A., Nelson, D. C. & Weijers, D. Evolution of plant hormone response pathways. Annu. Rev. Plant Biol. 71, 327–353 (2020).
Lew, T. T. S., Koman, V. B., Gordiichuk, P., Park, M. & Strano, M. S. The emergence of plant nanobionics and living plants as technology. Adv. Mater. Technol. 5, 1900657 (2020).
Giraldo, J. P., Wu, H., Newkirk, G. M. & Kruss, S. Nanobiotechnology approaches for engineering smart plant sensors. Nat. Nanotechnol. 14, 541–553 (2019).
Kwak, S. Y. et al. Nanosensor technology applied to living plant systems. Annu. Rev. Anal. Chem. 10, 113–140 (2017).
Zhu, C., Yang, G., Li, H., Du, D. & Lin, Y. Electrochemical sensors and biosensors based on nanomaterials and nanostructures. Anal. Chem. 87, 230–249 (2015).
Kwak, S. Y. et al. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 14, 447–455 (2019).
Lew, T. T. S. et al. Real-time detection of wound-induced H2O2 signalling waves in plants with optical nanosensors. Nat. Plants 6, 404–415 (2020).
Wong, M. H. et al. Nitroaromatic detection and infrared communication from wild-type plants using plant nanobionics. Nat. Mater. 16, 264–272 (2017).
Zrazhevskiy, P., Sena, M. & Gao, X. Designing multifunctional quantum dots for bioimaging, detection, and drug delivery. Chem. Soc. Rev. 39, 4326–4354 (2010).
Li, J., Wu, H., Santana, I., Fahlgren, M. & Giraldo, J. P. Standoff optical glucose sensing in photosynthetic organisms by a quantum dot fluorescent probe. ACS Appl. Mater. Interfaces 10, 28279–28289 (2018).
Li, W. et al. Phytotoxicity, uptake, and translocation of fluorescent carbon dots in mung bean plants. ACS Appl. Mater. Interfaces 8, 19939–19945 (2016).
Yao, Z., Lai, Z., Chen, C., Xiao, S. & Yang, P. Full-color emissive carbon-dots targeting cell walls of onion for in situ imaging of heavy metal pollution. Analyst 144, 3685 (2019).
Hasegawa, Y., Murohashi, F. & Uchida, H. Plant physiological activity sensing by bioelectric potential measurement. Procedia Engineer. 168, 630–633 (2016).
Ochiai, T., Tago, S., Hayashi, M. & Fujishima, A. Highly sensitive measurement of bio-electric potentials by boron-doped diamond (BDD) electrodes for plant monitoring. Sensors 15, 26921–26928 (2015).
Tenenboim, H. & Brotman, Y. Omic relief for the biotically stressed: metabolomics of plant biotic interactions. Trends Plant Sci. 21, 781–791 (2016).
Kushalappa, A. C. & Gunnaiah, R. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci. 18, 522–531 (2013).
Jorge, T. F. et al. Mass spectrometry-based plant metabolomics: metabolite responses to abiotic stress. Mass Spectrom. Rev. 35, 620–649 (2016).
Obata, T. & Fernie, A. R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 69, 3225–3243 (2012).
Ward, J. L. et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 63, 443–457 (2010).
Gunnaiah, R., Kushalappa, A. C., Duggavathi, R., Fox, S. & Somers, D. J. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat qtl (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE 7, e40695 (2012).
Kim, H. K., Choi, Y. H. & Verpoorte, R. NMR-based plant metabolomics: where do we stand, where do we go? Trends Biotechnol. 29, 267–275 (2011).
Cevallos-Cevallos, J. M., Futch, D. B., Shilts, T., Folimonova, S. Y. & Reyes-De-Corcuera, J. I. GC-MS metabolomic differentiation of selected citrus varieties with different sensitivity to citrus huanglongbing. Plant Physiol. Biochem. 53, 69–76 (2012).
Altangerel, N. et al. In vivo diagnostics of early abiotic plant stress response via Raman spectroscopy. Proc. Natl Acad. Sci. USA 114, 3393–3396 (2017).
Yeturu, S. et al. Handheld Raman spectroscopy for the early detection of plant diseases: Abutilon mosaic virus infecting Abutilon sp. Anal. Methods 8, 3450–3457 (2016).
Hao Huang, C. et al. Early diagnosis and management of nitrogen deficiency in plants utilizing Raman spectroscopy. Front. Plant Sci. 11, 663 (2020).
Farber, C., Mahnke, M., Sanchez, L. & Kurouski, D. Advanced spectroscopic techniques for plant disease diagnostics. A review. Trends Analyt. Chem. 118, 43–49 (2019).
Angel, S. M., Gomer, N. R., Sharma, S. K. & McKay, C. Remote Raman spectroscopy for planetary exploration: a review. Appl. Spectrosc. 66, 137–50 (2012).
Acosta-Maeda, T. E. et al. Remote Raman measurements of minerals, organics, and inorganics at 430 m range. Appl. Opt. 55, 10283 (2016).
Dudareva, N., Negre, F., Nagegowda, D. A. & Orlova, I. Plant volatiles: recent advances and future perspectives. CRC. Crit. Rev. Plant Sci. 25, 417–440 (2006).
Vivaldo, G., Masi, E., Taiti, C., Caldarelli, G. & Mancuso, S. The network of plants volatile organic compounds. Sci. Rep. 7, 11050 (2017).
Martinelli, F. et al. Advanced methods of plant disease detection. A review. Agro. Sustain. Dev. 35, 1–25 (2015).
Tholl, D. et al. Practical approaches to plant volatile analysis. Plant J. 45, 540–560 (2006).
Li, Z. et al. Non-invasive plant disease diagnostics enabled by smartphone-based fingerprinting of leaf volatiles. Nat. Plants 5, 856–866 (2019).
Fong, D., Luo, S. X., Andre, R. S. & Swager, T. M. Trace ethylene sensing via wacker oxidation. ACS Cent. Sci. 6, 507–512 (2020).
Cui, S., Ling, P., Zhu, H. & Keener, H. Plant pest detection using an artificial nose system: a review. Sensors 18, 378 (2018).
Zhang, Y. et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 667–697 (2016).
Chen, K., Wang, Y., Zhang, R., Zhang, H. & Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697 (2019).
Singh, A. K., Ganapathysubramanian, B., Sarkar, S. & Singh, A. Deep learning for plant stress phenotyping: trends and future perspectives. Trends Plant Sci. 23, 883–898 (2018).
Fichman, Y., Miller, G. & Mittler, R. Whole-plant live imaging of reactive oxygen species. Mol. Plant 12, 1203–1210 (2019).
Dhondt, S., Wuyts, N. & Inzé, D. Cell to whole-plant phenotyping: the best is yet to come. Trends Plant Sci. 18, 428–439 (2013).
Giraldo, J. P. et al. Plant nanobionics approach to augment photosynthesis and biochemical sensing. Nat. Mater. 13, 400–408 (2014).
Cunningham, F. J., Goh, N. S., Demirer, G. S., Matos, J. L. & Landry, M. P. Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897 (2018).
Lew, T. T. S. et al. Rational design principles for the transport and subcellular distribution of nanomaterials into plant protoplasts. Small 14, 1802086 (2018).
Lew, T. T. S. et al. Nanocarriers for transgene expression in pollen as a plant biotechnology tool. ACS Mater. Lett. 2, 1057–1066 (2020).
Thagun, C., Chuah, J. & Numata, K. Targeted gene delivery into various plastids mediated by clustered cell‐penetrating and chloroplast‐targeting peptides. Adv. Sci. 6, 1902064 (2019).
Yoshizumi, T., Oikawa, K., Chuah, J. A., Kodama, Y. & Numata, K. Selective gene delivery for integrating exogenous DNA into plastid and mitochondrial genomes using peptide-DNA complexes. Biomacromolecules 19, 1582–1591 (2018).
Hadjidemetriou, M. & Kostarelos, K. Nanomedicine: evolution of the nanoparticle corona. Nat. Nanotechnol. 12, 288–290 (2017).
Zhdanov, V. P. Formation of a protein corona around nanoparticles. Curr. Opin. Colloid Interface Sci. 41, 95–103 (2019).
Ke, P. C., Lin, S., Parak, W. J., Davis, T. P. & Caruso, F. A decade of the protein corona. ACS Nano 11, 11773–11776 (2017).
Cai, R. et al. Corona of thorns: the surface chemistry-,ediated protein corona perturbs the recognition and immune response of macrophages. ACS Appl. Mater. Interfaces 12, 1997–2008 (2020).
Thapa, S. & Prasanna, R. Prospecting the characteristics and significance of the phyllosphere microbiome. Ann. Microbiol. 68, 229–245 (2018).
Mohanram, S. & Kumar, P. Rhizosphere microbiome: revisiting the synergy of plant-microbe interactions. Ann. Microbiol. 69, 307–320 (2019).
Bai, Y. et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015).
Hassani, M. A., Durán, P. & Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 6, 58 (2018).
Busby, P. E. et al. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 15, e2001793 (2017).
Müller, D. B., Vogel, C., Bai, Y. & Vorholt, J. A. The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50, 211–234 (2016).
Kozawa, D. et al. A fiber optic interface coupled to nanosensors: applications to protein aggregation and organic molecule guantification. ACS Nano 14, 10141–10152 (2020).
Di, W. & Clark, H. A. Optical nanosensors for: in vivo physiological chloride detection for monitoring cystic fibrosis treatment. Anal. Methods 12, 1441–1448 (2020).
Parikh, S. J., Goyne, K. W., Margenot, A. J., Mukome, F. N. D. & Calderón, F. J. in Advances in Agronomy Vol. 126 (Academic Press Inc., 2014).
Wasson, A., Bischof, L., Zwart, A. & Watt, M. A portable fluorescence spectroscopy imaging system for automated root phenotyping in soil cores in the field. J. Exp. Bot. 67, 1033–1043 (2016).
Tracy, S. R. et al. Crop Improvement from phenotyping roots: highlights reveal expanding opportunities. Trends Plant Sci. 25, 105–118 (2020).
Gu, K. et al. Crosskingdom growth benefits of fungus-derived phytohormones in Choy Sum. Preprint at bioRxiv https://doi.org/10.1101/2020.02.04.933770 (2020).
Polisetti, S., Bible, A. N., Morrell-Falvey, J. L. & Bohn, P. W. Raman chemical imaging of the rhizosphere bacterium Pantoea sp. YR343 and its co-culture with Arabidopsis thaliana. Analyst 141, 2175–2182 (2016).
Singer, E., Wagner, M. & Woyke, T. Capturing the genetic makeup of the active microbiome in situ. ISME J. 11, 1949–1963 (2017).
Gehan, M. A. et al. PlantCV v2: image analysis software for high-throughput plant phenotyping. PeerJ 5, e4088 (2017).
Rodriguez, P. A. et al. Systems biology of plant–microbiome interactions. Mol. Plant 12, 804–821 (2019).
Toju, H. et al. Core microbiomes for sustainable agroecosystems. Nat. Plants 4, 247–257 (2018).
Ballaré, C. L. & Pierik, R. The shade-avoidance syndrome: multiple signals and ecological consequences. Plant. Cell Environ. 40, 2530–2543 (2017).
Tang, Y.-J. & Liesche, J. The molecular mechanism of shade avoidance in crops — how data from Arabidopsis can help to identify targets for increasing yield and biomass production. J. Integr. Agr. 16, 1244–1255 (2017).
Wille, W., Pipper, C. B., Rosenqvist, E., Andersen, S. B. & Weiner, J. Reducing shade avoidance responses in a cereal crop. AoB Plants 9, plx039 (2017).
Casal, J. J. Shade avoidance. Arab. B. 10, e0157 (2012).
Devlin, P. F., Yanovsky, M. J. & Kay, S. A. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133, 1617–1629 (2003).
Rolauffs, S., Fackendahl, P., Sahm, J., Fiene, G. & Hoecker, U. Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Physiol. 160, 2015–2027 (2012).
Asseng, S. et al. Rising temperatures reduce global wheat production. Nat. Clim. Change 5, 143–147 (2015).
Pereira, A. Plant abiotic stress challenges from the changing environment. Front. Plant Sci. 7, 1123 (2016).
Elad, Y. & Pertot, I. Climate change impacts on plant pathogens and plant diseases. J. Crop Improv. 28, 99–139 (2014).
Piao, S. et al. The impacts of climate change on water resources and agriculture in China. Nature 467, 43–51 (2010).
Biermacher, J. T., Brorsen, B. W., Epplin, F. M., Solie, J. B. & Raun, W. R. The economic potential of precision nitrogen application with wheat based on plant sensing. Agric. Econ. 40, 397–407 (2009).
Polo, E. & Kruss, S. Impact of redox-active molecules on the fluorescence of polymer-wrapped carbon nanotubes. J. Phys. Chem. C 120, 3061–3070 (2016).
Wang, L., Luo, W., Sun, X. & Qian, C. Changes in flavor-relevant compounds during vine ripening of tomato fruit and their relationship with ethylene production. Hortic. Environ. Biotechnol. 59, 787–804 (2018).
Klee, H. J. & Giovannoni, J. J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59 (2011).
Akhatou, I. & Fernández-Recamales, Á. Nutritional and nutraceutical quality of strawberries in relation to harvest time and crop conditions. J. Agric. Food Chem. 62, 5749–5760 (2014).
Lowry, G. V., Avellan, A. & Gilbertson, L. M. Opportunities and challenges for nanotechnology in the agri-tech revolution. Nat. Nanotechnol. 14, 517–522 (2019).
Kah, M., Kookana, R. S., Gogos, A. & Bucheli, T. D. A critical evaluation of nanopesticides and nanofertilizers against their conventional analogues. Nat. Nanotechnol. 13, 677–684 (2018).
Falinski, M. M. et al. A framework for sustainable nanomaterial selection and design based on performance, hazard, and economic considerations. Nat. Nanotechnol. 13, 708–714 (2018).
Hofmann, T. et al. Technology readiness and overcoming barriers to sustainably implement nanotechnology-enabled plant agriculture. Nat. Food 1, 416–425 (2020).
Eschen, R. et al. Safeguarding global plant health: the rise of sentinels. J. Pest Sci. 92, 29–36 (2019).
Kenis, M. et al. Sentinel nurseries to assess the phytosanitary risks from insect pests on importations of live plants. Sci. Rep. 8, 11217 (2018).
Radoglou-Grammatikis, P., Sarigiannidis, P., Lagkas, T. & Moscholios, I. A compilation of UAV applications for precision agriculture. Comput. Networks 172, 107148 (2020).
Santesteban, L. G. et al. High-resolution UAV-based thermal imaging to estimate the instantaneous and seasonal variability of plant water status within a vineyard. Agric. Water Manag. 183, 49–59 (2017).
Saiz-Rubio, V. & Rovira-Más, F. From smart farming towards agriculture 5.0: a review on crop data management. Agronomy 10, 207 (2020).
Acknowledgements
The authors are grateful for the funding and support from the MIT-Singapore SMART programme, specifically the Disruptive and Sustainable Technology for Agricultural Precision (DiSTAP) integrated research group, and the National Research Foundation, Singapore (NRF-CRP16-2015-04 to N.I.N). T.T.S.L. was supported on a graduate fellowship by the Agency of Science, Research and Technology, Singapore. We thank B. Skrip (Massachusetts Institute of Technology) for creating Fig. 2 in this manuscript.
Author information
Authors and Affiliations
Contributions
T.T.S.L., N.-H.C. and M.S.S. led the writing of this manuscript. T.T.S.L., R.S., I.-C.J., B.S.P., N.I.N., M.H.W., G.P.S., R.J.R., O.S., K.S., N.-H.C. and M.S.S. contributed to the writing of specific sections, critical reading of the manuscript and the reviewing of appropriate references.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lew, T.T.S., Sarojam, R., Jang, IC. et al. Species-independent analytical tools for next-generation agriculture. Nat. Plants 6, 1408–1417 (2020). https://doi.org/10.1038/s41477-020-00808-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-020-00808-7
This article is cited by
-
Bioelectronics for basic plant science and precision agriculture
Nature Reviews Electrical Engineering (2026)
-
Accelerating adoption of species-agnostic plant sensors for precision farming
Nature Reviews Electrical Engineering (2025)
-
Nanofabrication of silk microneedles for high-throughput micronutrient delivery and continuous sap monitoring in plants
Nature Nanotechnology (2025)
-
RETRACTED ARTICLE: Harnessing NMR technology for enhancing field crop improvement: applications, challenges, and future perspectives
Metabolomics (2025)
-
Durable, recyclable, and stretchable plasmonic film for grapefruit decay detection
Analytical and Bioanalytical Chemistry (2025)