Abstract
Phosphorylation of histone H3 at threonine 11 (H3T11ph) affects transcription and chromosome stability. However, the enzymes responsible for depositing H3T11ph and the functions of H3T11ph in plants remain unknown. Here we report that in Arabidopsis thaliana, PYRUVATE KINASE 6 (PK6), PK7 and PK8 enter the nucleus under conditions of sufficient glucose and light exposure, where they interact with SWI2/SNF2-RELATED 1 COMPLEX 4 (SWC4) and phosphorylate H3 at threonine 11. Mutations in these kinases or knockdown of SWC4 resulted in FLC-dependent early flowering, short hypocotyls and short pedicels. Genome-wide, H3T11ph is highly enriched at transcription start sites and transcription termination sites, and positively correlated with gene transcript levels. PK6 and SWC4 targeted FLC, MYB73, PRE1, TCP4 and TCP10, depositing H3T11ph at these loci and promoting their transcription, and PK6 occupancy at these loci requires SWC4. Together, our results reveal that nuclear-localized PK6, PK7 and PK8 modulate H3T11ph and plant growth.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The Arabidopsis mutants used in the current study are available from the corresponding author on request. The RNA-seq data and ChIP-seq data were deposited to the NCBI Sequence Read Archive with accession code PRJNA1122224. The source data underlying Figs. 1a,b, 2e,h,i, 4a and 5b,e–g; Extended Data Figs. 4a, 6d–f and 8j,k; and Supplementary Figs. 1, 3d and 8a–c are provided as a Source Data file. Other source data are provided with this paper.
References
Fischle, W. et al. Regulation of HP1–chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005).
Komar, D. & Juszczynski, P. Rebelled epigenome: histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy. Clin. Epigenetics 12, 147 (2020).
Lau, P. N. I. & Cheung, P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc. Natl Acad. Sci. USA 108, 2801–2806 (2011).
Yang, W. et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685–696 (2012).
Xu, D., Liang, J., Lin, J. & Yu, C. PKM2: a potential regulator of rheumatoid arthritis via glycolytic and non-glycolytic pathways. Front. Immunol. 10, 2919 (2019).
Zhang, Z. et al. PKM2, function and expression and regulation. Cell Biosci. 9, 52 (2019).
Dombrauckas, J. D., Santarsiero, B. D. & Mesecar, A. D. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry 44, 9417–9429 (2005).
Gao, X., Wang, H., Yang, J. J., Liu, X. & Liu, Z.-R. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 45, 598–609 (2012).
Wang, G. & Han, J. J. Connections between metabolism and epigenetic modifications in cancer. Med. Rev. 1, 199–221 (2021).
Li, S. et al. Serine and SAM responsive complex SESAME regulates histone modification crosstalk by sensing cellular metabolism. Mol. Cell 60, 408–421 (2015).
He, F. et al. SESAME-catalyzed H3T11 phosphorylation inhibits Dot1-catalyzed H3K79me3 to regulate autophagy and telomere silencing. Nat. Commun. 13, 7526 (2022).
Andre, C., Froehlich, J. E., Moll, M. R. & Benning, C. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19, 2006–2022 (2007).
Baud, S. et al. Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J. 52, 405–419 (2007).
Hu, L. et al. Mitochondria-associated pyruvate kinase complexes regulate grain filling in rice. Plant Physiol. 183, 1073–1087 (2020).
Oliver, S. N. et al. Decreased expression of cytosolic pyruvate kinase in potato tubers leads to a decline in pyruvate resulting in an in vivo repression of the alternative oxidase. Plant Physiol. 148, 1640–1654 (2008).
Knowles, V. L. et al. Altered growth of transgenic tobacco lacking leaf cytosolic pyruvate kinase. Plant Physiol. 116, 45–51 (1998).
Zhang, B. & Liu, J.-Y. Cotton cytosolic pyruvate kinase GhPK6 participates in fast fiber elongation regulation in a ROS-mediated manner. Planta 244, 915–926 (2016).
Michaels, S. D. & Amasino, R. M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999).
Choi, K. et al. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134, 1931–1941(2007).
Lázaro, A., Gomez-Zambrano, A., López-González, L., Pineiro, M. & Jarillo, J. A. Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 59, 653–666 (2008).
Noh, Y.-S. & Amasino, R. M. PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15, 1671–1682 (2003).
Gómez-Zambrano, Á. et al. Arabidopsis SWC4 binds DNA and recruits the SWR1 complex to modulate histone H2A.Z deposition at key regulatory genes. Mol. Plant 11, 815–832 (2018).
Nguyen, V. Q. et al. Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1. Cell 154, 1220–1231 (2013).
Su, Y. et al. Phosphorylation of histone H2A at serine 95: a plant-specific mark involved in flowering time regulation and H2A.Z deposition. Plant Cell 29, 2197–2213 (2017).
Yi, H. et al. Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell 18, 1575–1589 (2006).
Xiong, Y. et al. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186 (2013).
Ye, R. et al. Glucose-driven TOR–FIE–PRC2 signalling controls plant development. Nature 609, 986–993 (2022).
Shi, L., Wu, Y. & Sheen, J. TOR signaling in plants: conservation and innovation. Development 145, dev160887 (2018).
Fu, L. et al. The TOR–EIN2 axis mediates nuclear signalling to modulate plant growth. Nature 591, 288–292 (2021).
Gibon, Y. et al. Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post‐translational activation of ADP‐glucose pyrophosphorylase in the following light period. Plant J. 39, 847–862 (2004).
Zhang, S. et al. Metabolic regulation of telomere silencing by SESAME complex-catalyzed H3T11 phosphorylation. Nat. Commun. 12, 594 (2021).
Gu, X. et al. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4, 1947 (2013).
Teper-Bamnolker, P. & Samach, A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17, 2661–2675 (2005).
Yang, Y. et al. UV‐B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J. 39, e101928 (2020).
Challa, K. R., Aggarwal, P. & Nath, U. Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28, 2117–2130 (2016).
Cai, H. et al. ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin‐mediated activation of PRE 1 expression. New Phytol. 214, 1579–1596 (2017).
Ikram, A. U. et al. Chromatin remodeling factors OsYAF9 and OsSWC4 interact to promote internode elongation in rice. Plant Physiol. 188, 2199–2214 (2022).
Rolland, F., Moore, B. & Sheen, J. Sugar sensing and signaling in plants. Plant Cell 14, S185–S205 (2002).
Mason, M. G., Ross, J. J., Babst, B. A., Wienclaw, B. N. & Beveridge, C. A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl Acad. Sci. USA 111, 6092–6097 (2014).
Rosa, M. et al. Soluble sugars: metabolism, sensing and abiotic stress: a complex network in the life of plants. Plant Signal. Behav. 4, 388–393 (2009).
Ohto, M. et al. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 127, 252–261 (2001).
Zhou, L., Jang, J.-C., Jones, T. L. & Sheen, J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl Acad. Sci. USA 95, 10294–10299 (1998).
Niu, Y. et al. HISTONE DEACETYLASE 9 transduces heat signal in plant cells. Proc. Natl Acad. Sci. USA 119, e2206846119 (2022).
Zhu, D. et al. Distinct chromatin signatures in the Arabidopsis male gametophyte. Nat. Genet. 55, 706–720 (2023).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32230017, U19A2021 and 32321001), the Global Select Project (DJK-LX-2022007) of the Institute of Health and Medicine, the Hefei Comprehensive National Science Center, the Key Research and Development Projects in Anhui Province (2023n06020035), and the Fundamental Research Funds for the Central Universities (WK9100000047). We thank Y. Xiong from the FuJian Agriculture and Forestry University for kindly providing the tor-es and HA-S6K-overexpressing line seeds, and all members of the Ding group for helpful discussions.
Author information
Authors and Affiliations
Contributions
Y.D. and P.H. conceived the study and designed the experiments. P.H. performed most of the experiments. Y.D. wrote the manuscript. Y. Xu, Y.S. and Y.W. contributed to mutant generation, and Y. Xiong contributed to glucose starvation design.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Tamaki Suganuma, Keqiang Wu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Reproducibility analysis of the biological replicates for the H3T11ph ChIP-seq.
(a) Correlation analysis of the biological replicates of the H3T11ph ChIP-seq analysis of Col-0 under glucose depletion and stimulation. The x and y axes indicate the read coverage normalized to the total mapped reads, and values are in the log2 scale. (b) Scatterplot of H3T11ph-occupied loci downregulated in Col-0 under glucose depletion vs. glucose stimulation. The x-axis indicates log2-normalized ChIP read counts, and the y-axis indicates the log2 fold change of H3T11ph levels. The dashed line indicates a fold change of 1.5. Significant (FDR < 0.05) up- and downregulated peaks are marked in red and blue, respectively. The up- and downregulated loci are indicated. (c) Metagene plots of H3T11ph enrichment in Col-0 under glucose depletion and stimulation. The log2 ratio of normalized enrichment (H3T11ph/Input) for each gene region, including the 2-kbp upstream and downstream regions from the gene, was used for the average gene analysis. The transcription start site (TSS) and transcription termination site (TTS) are indicated. (d) ChIP–seq for H3T11ph in Col-0 under glucose depletion and stimulation. The genome-browser track views are shown (chr3: 9,391,000–9,638,000 and chr4: 14,036,000–14,302,000).
Extended Data Fig. 2 Phylogenetic analysis of pyruvate kinase (PK) family proteins in Arabidopsis.
The 14 PKs, including 10 cytosolic PKs and 4 plastidic PKs in Arabidopsis were aligned with ClustalW. The relationships of the sequences were examined with MEGA5. The evolutional scale bar is indicated at the bottom.
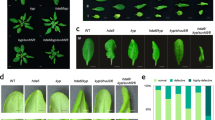
Extended Data Fig. 3 Characterization of the pk1, pk2, pk3, pk4, pk5, pk6, pk7, pk8, pk9, and pk10 mutants.
(a) Gene structures of PK1, PK2, PK3, PK4, PK5, PK6, PK7, PK8, PK9, and PK10 with boxes indicating exons, lines indicating introns, and triangles indicating T-DNA insertions. The primers used for genotyping are marked with arrows. (b) Genotypic analysis of the pk1, pk2, pk3, pk4, pk5, pk6, pk7, pk8, pk9, and pk10 mutants. Genotyping was performed with the left genomic primer (LP), right genomic primer (RP), and the vector primer (LBb1.3). The positions of the LPs and RPs are indicated in (A). (c) Expression levels of the full-length transcripts of PK1, PK2, PK3, PK4, PK5, PK6, PK7, PK8, PK9, and PK10 in the pk1, pk2, pk3, pk4, pk5, pk6, pk7, pk8, pk9, and pk10 mutants, respectively.
Extended Data Fig. 4 Phosphorylation activity of PK6.
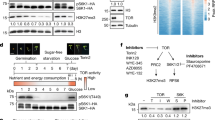
(a) Global H3T11ph levels in pk1-1 pk2-1, pk2-1 pk3-1 pk4-1, pk3-1 pk4-1 pk5-1, pk9-1 pk10-1, and pk6-1 pk7-1 pk8-1 plants. H3 was used as a loading control. Replicate 1 and replicate 2 are shown. All experiments were repeated at least three times, and representative experiments are shown. Molecular mass markers in kilodaltons (kDa) are indicated on the left, and the bands are consistent with their expected sizes. The source data underlying immunoblots are provided in Source Data File. (b) Phosphorylation activity of PK6 purified from Escherichia coli on H3. SnrK2.6 was used as the positive control. The positions of MBP-PK6, GST-SnrK2.6, and H3 are marked with arrows. The star indicates the no-specific band. (c) Kinase activity of immunoprecipitated PK6 on the histones. The positions of H2A, H2B, H3, and H4 are marked with arrows. (d) The conserved arginine 87 and lysine 172 of PK6 were converted to glutamic acid and arginine, respectively (PK6R87E/K172R), to investigate the requirement of these conserved residues for the phosphorylation of Arabidopsis H3 in vitro. The conserved ATP binding sites are shown in Supplemental Fig. 7a. GFP levels were used as a loading control. (e) Activity and specificity of the immunoprecipitated PK6 kinase, assessed using different substrates. The substrates were H3 fused with His and either wild-type H3 or H3 with serine-to-alanine substitutions. GFP levels were used as a loading control.
Extended Data Fig. 5 The GUS activity of PK6 under glucose depletion and stimulation.
(a) The PK6-driven GUS activity was assessed under glucose depletion and stimulation in four-day-old seedlings. Bar, 1 mm. (b) Relative transcript levels of PK6, PK7, and PK8 under glucose depletion and stimulation. Experiments were repeated at least three times, and the results of representative experiments are shown as means ± SE, n = 3. No difference is indicated as ns. Tubulin 8 was used as a negative control. The source data underlying the statistical analysis are provided in the Source Data file.
Extended Data Fig. 6 Phosphorylation activity of H3 at threonine 11 by PK7 and PK8.
(a) Complementation of the pk7-1 and pk8-1 mutants with full-length PK7 or PK8 sequences, respectively, fused with HA driven by the PK7 (ProPK7:HA-PK7) or PK8 (ProPK8:HA-PK8) promoters, respectively. Over 40 independent transgenic lines were generated for each transformant, and two representative transgenic plants (T7 and T10 for the pk7-1 complementation, and T2 and T8 for the pk8-1 complementation) are shown. (b) Total leaf number of Col-0, pk7-1, pk8-1, and the complemented lines under a long-day photoperiod. Flowering time was assessed by counting the number of rosette leaves (white boxes) and cauline leaves (grey boxes) at bolting under a long-day photoperiod. Values shown are means ± SD of total leaves; over 25 plants were scored for each line. The source data underlying the statistical analysis are provided in the Source Data file. (c) Activity of the immunoprecipitated PK7 (left panel) and PK8 (right panel) kinases on H3 under glucose depletion and stimulation. Experiments were repeated at least three times and the results of representative experiments are shown. (d) Activity of the immunoprecipitated PK7 and PK8 kinases on threonine 11 of H3 under glucose stimulation. Experiments were repeated at least three times and the results of representative experiments are shown. The source data underlying immunoblots are provided in Source Data File. (e) The total PK7 levels and PK7 in the nucleus and cytoplasm of ProPK7:HA-PK7 plants (T7) under glucose depletion and stimulation, determined using a western blot. A total of 4% nuclear proteins and 20% cytoplasmic proteins were used for western blotting. The nuclear and cytoplasmic proteins were validated by anti-H3 and anti-β-Tublin, respectively. (f) The total PK8 levels and PK8 levels in the nucleus and cytoplasm of ProPK8:HA-PK8 plants (T2) under glucose depletion and stimulation, determined using a western blot. The nuclear and cytoplasmic proteins were validated by anti-H3 and anti-β-Tubulin, respectively. The source data underlying immunoblots in (E) and (F) are provided in Source Data File.
Extended Data Fig. 7 The PK activity of PK6, PK7, and PK8.
(a) Sequence alignment of PK6 with PKM2 and Pyk1. The conserved ATP binding sites are shown in red boxes, and PEP binding sites are shown in green boxes. (b) PK6, PK7, PK8, and PK6 with conserved arginine 46 and lysine 236 converted to glutamic acid and arginine, respectively (PK6R46E/K236R), as shown in a Coomassie blue-stained gel. (c) The PK activity of PK6, PK7, PK8, and PK6R46E/K236R. Values are means ± SD (n = 3 independent biological replicates) (d) PK activity in Col-0 and pk6/7/8 seedlings under glucose depletion and glucose stimulation. Values are means ± SD (n = 3 independent biological replicates). (e) Glucose levels in Col-0 under dark treatment and LD treatment. Values are means ± SD (n = 3 independent biological replicates). The source data underlying the statistical analysis in (c), (d) and (e) are provided in the Source Data file.
Extended Data Fig. 8 The translocation of PK6, PK7, and PK8 under glucose stimulation and depletion.
(a), (c), (e), and (h) Cellular sublocations of PK6-GFP (a), PK7-GFP (c), PK8-GFP (e), and GFP alone (h) in Arabidopsis protoplasts under glucose stimulation and glucose plus AZD8055 or Torin2. H3-RFP was used as a nuclear marker. GFP, H3-RFP, bright-field microscopy (differential interference contrast; DIC), and merged images are shown. Scale bars, 10 µm. GFP alone (h) was used as the negative control. GFP, green fluorescent protein. RFP, red fluorescent protein. (b), (d), (f), and (i) Quantitative confocal imaging of PK6-GFP (b), PK7-GFP (d), PK8-GFP (f), and GFP alone (i) in Arabidopsis protoplasts, determined by calculating the cytoplasm/nucleus (C/N) signal intensity ratio. Values shown are means ± SD; over 10 cells were scored for each treatment. Different letters mark statistically significant differences, as revealed using a one-way ANOVA (p < 0.01). (j) and (k) H3T11ph in Col-0 and PK6 levels in 35S:GFP-PK6 plants were measured during a darkness plus glucose and re-light treatment (j), and constituted long day plus glucose treatment (k) using a western blot. Molecular mass markers in kilodaltons (kDa) are indicated on the left in, and the bands are consistent with their expected sizes. H3 was used as a loading control. Experiments were repeated at least three times, and the results of representative experiments are shown. The source data underlying immunoblots are provided in Source Data File. (l) Relative transcription levels of PK6, PK7, and PK8 under long day photoperiod, darkness, and re-light conditions. Experiments were repeated at least three times. and the results of representative experiments are shown as means ± SE, n = 3. Ubiquitin 10 was used as internal control. The source data underlying the statistical analysis in (b), (d), (f), (i) and (l) are provided in the Source Data file.
Extended Data Fig. 9 Characterization of the pk6 mutants.
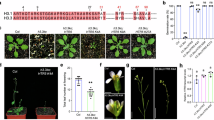
(a) Total leaf number of pk6-1, pk6-2, pk7-1, pk8-1, and pk6/7/8 mutants under a long-day photoperiod, a short-day photoperiod, and a short-day photoperiod with gibberellic acid (GA3) treatment. White boxes indicate rosette leaves and grey boxes indicate cauline leaves. Values shown are means ± SD of total leaves; over 25 plants were scored for each line. (b) Flowering-time phenotypes of Col-0, pk6-1, and two complemented lines (T16 and T18) under a long-day photoperiod. Over 40 independent transgenic lines were generated, and two representative transgenic plants are shown. (c) Total leaf number of Col-0, pk6-1, and two complemented lines under a long-day photoperiod. Flowering time was assessed by counting the number of rosette leaves and cauline leaves at bolting under the long-day photoperiod. White boxes indicate rosette leaves, and grey boxes indicate cauline leaves. Values shown are means ± SD of total leaves; over 25 plants were scored for each line. (d) Representative images of FRI, FRI pk6-1, FRI pk6-2, FRI pk7-1, FRI pk8-1, and FRI pk6/7/8 plants under a long-day photoperiod. (e) Total leaf number of FRI, FRI pk6-1, FRI pk6-2, FRI pk7-1, FRI pk8-1, and FRI pk6/7/8 plants under a long-day photoperiod. White boxes indicate rosette leaves, and grey boxes indicate cauline leaves. Values shown are means ± SD of total leaves; over 25 plants were scored for each line. (f) and (g) Relative transcript levels of FLC (f) and FT (g) in FRI pk6-1, FRI pk6-2, FRI pk7-1, FRI pk8-1, and FRI pk6/7/8. Experiments were repeated at least three times and the results of representative experiments are shown as means ± SE, n = 3. Asterisks indicate a significant difference using Student’s t-test (P < 0.01). No difference is indicated as ns. Ubiquitin 10 was used as internal control. The source data underlying the statistical analysis in (a), (c), (e), (f) and (g) are provided in the Source Data file.
Extended Data Fig. 10 Reproducibility analysis of the biological replicates for the RNA-seq.
(a) and (b) Correlation analysis of the biological replicates of the RNA-seq analysis of Col-0 (a) and pk6/7/8 (b) under a long-day photoperiod. The x and y axes indicate the read coverage normalized to the total mapped reads, and values are in the log2 scale. (c) Volcano plot of the RNA-seq results of Col-0 and pk6/7/8. The negative log of the p-value (base 10) was plotted on the y axis, while the log of the fold change (FC; base 2) was plotted on the x axis. The upregulated and downregulated genes in pk6/7/8 compared with Col-0 are indicated in the Venn diagram. (d) Gene ontology (GO) enrichment of functions associated with biological processes in the 2355 downregulated genes in pk6/7/8. The differentially expressed genes were analysed using TBtools (v.2.052). The p-value in Fisher exact test is computed.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and Source Data for unprocessed western blots in supplementary figures.
Supplementary Table 1
H3T11ph peaks with or without glucose.
Supplementary Table 2
Upregulated and downregulated genes in pk6/7/8 mutants.
Supplementary Table 3
H3T11ph peaks in Col-0 and pk6/7/8 plants.
Supplemental Dataset 1
The primers and constructs.
Supplemental Dataset 2
Statistical source data for supplementary figures.
Source data
Source Data Fig. 1
Unprocessed western blots for Figs. 1a,b,d.
Source Data Fig. 2
Unprocessed western blots for Fig. 2e.
Source Data Fig. 2–4, 7 and 8, and Source Data Extended Data Figs. 5–9
Statistical source data for Fig. 2d. Statistical source data for Fig. 3b. Statistical source data for Fig. 4b,d,f,g,j. Statistical source data for Fig. 7f,g,i,j. Statistical source data for Fig. 8b–d. Statistical source data for Extended Data Fig. 5b. Statistical source data for Extended Data Fig. 6b. Statistical source data for Extended Data Fig. 7c–e. Statistical source data for Extended Data Fig. 8b,d,f,i,l. Statistical source data for Extended Data Fig. 9a,c,e–g.
Source Data Fig. 3
Unprocessed western blots for Fig. 3c,d.
Source Data Fig. 5
Unprocessed western blots for Fig. 5a.
Source Data Fig. 6
Unprocessed western blots for Fig. 6b,e–g.
Source Data Extended Data Fig. 4
Unprocessed western blots for Extended Data Fig. 4a.
Source Data Extended Data Fig. 6
Unprocessed western blots for Extended Data Fig. 6d–f.
Source Data Extended Data Fig. 8
Unprocessed western blots for Extended Data Fig. 8j,k.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, P., Xu, Y., Su, Y. et al. Nuclear-localized pyruvate kinases control phosphorylation of histone H3 on threonine 11. Nat. Plants 10, 1682–1697 (2024). https://doi.org/10.1038/s41477-024-01821-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-024-01821-w