Abstract
In flowering plants, rapid activation of the zygotic genome occurs after fertilization1,2,3, but there is limited knowledge of the molecular pathways underlying embryo initiation4. In rice, a key role is played by the transcription factor BABY BOOM 1 (OsBBM1), initially expressed from the paternal genome1. Ectopic OsBBM1 expression in the egg cell can override the fertilization requirement, giving rise to parthenogenetic progeny5. Here we show that the WOX-family transcription factor DWARF TILLER1 (OsDWT1)/WUSCHEL-LIKE HOMEODOMAIN 9 (OsWOX9A)6, another gene paternally expressed in zygotes, is a strong enhancer of embryo initiation by OsBBM1. Co-expression of OsWOX9A and OsBBM1 in egg cells results in 86–91% parthenogenesis, representing 4- to 15-fold increases over OsBBM1 alone. These results suggest that embryo initiation is promoted by the synergistic action of paternal-genome-expressed transcription factors in the fertilized egg cell. These findings can be utilized for the efficient production of haploids, as well as clonal hybrid seeds in crop plants7,8.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. The genetic materials are available from the corresponding authors upon request. The rice reference genome sequence is publicly available at http://rice.uga.edu/. Source data are provided with this paper.
References
Anderson, S. N. et al. The zygotic transition is initiated in unicellular plant zygotes with asymmetric activation of parental genomes. Dev. Cell 43, 349–358.e4 (2017).
Chen, J. et al. Zygotic genome activation occurs shortly after fertilization in maize. Plant Cell 29, 2106–2125 (2017).
Zhao, P. et al. Two-step maternal-to-zygotic transition with two-phase parental genome contributions. Dev. Cell 49, 882–893.e5 (2019).
Zhao, P., Shi, C., Wang, L. & Sun, M. The parental contributions to early plant embryogenesis and the concept of maternal-to-zygotic transition in plants. Curr. Opin. Plant Biol. 65, 102144 (2022).
Khanday, I., Skinner, D., Yang, B., Mercier, R. & Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95 (2019).
Wang, W. et al. DWARF TILLER1, a WUSCHEL-related homeobox transcription factor, is required for tiller growth in rice. PLoS Genet. 10, e1004154 (2014).
Xiong, J. et al. Synthetic apomixis: the beginning of a new era. Curr. Opin. Biotechnol. 79, 102877 (2023).
Underwood, C. J. & Mercier, R. Engineering apomixis: clonal seeds approaching the fields. Annu. Rev. Plant Biol. 73, 201–225 (2022).
Anderson, S. N. et al. Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 76, 729–741 (2013).
Kim, S., Soltis, P. S., Wall, K. & Soltis, D. E. Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 23, 107–120 (2006).
Steffen, J. G., Kang, I.-H., Macfarlane, J. & Drews, G. N. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51, 281–292 (2007).
Stroud, H. et al. Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2, e00354 (2013).
Vernet, A. et al. High-frequency synthetic apomixis in hybrid rice. Nat. Commun. 13, 7963 (2022).
Skinner, D. J., Mallari, M. D., Zafar, K., Cho, M.-J. & Sundaresan, V. Efficient parthenogenesis via egg cell expression of maize BABY BOOM 1: a step toward synthetic apomixis. Plant Physiol. https://doi.org/10.1093/plphys/kiad461 (2023).
Sakai, H. et al. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54, e6 (2013).
Kawahara, Y. et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4 (2013).
Ouyang, S. et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 35, D883–D887 (2007).
Fang, F., Ye, S., Tang, J., Bennett, M. J. & Liang, W. DWT1/DWL2 act together with OsPIP5K1 to regulate plant uniform growth in rice. New Phytol. 225, 1234–1246 (2020).
Wu, X., Dabi, T. & Weigel, D. Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr. Biol. 15, 436–440 (2005).
Breuninger, H., Rikirsch, E., Hermann, M., Ueda, M. & Laux, T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14, 867–876 (2008).
Doudna, J. A. & Charpentier, E. The new frontier of genome engineering with CRISPR–Cas9. Science 346, 1258096 (2014).
Tang, J., Huang, X., Sun, M. & Liang, W. DWARF TILLER1 regulates apical–basal pattern formation and proper orientation of rice embryos. Plant Physiol. https://doi.org/10.1093/plphys/kiae318 (2024).
Armenta-Medina, A. & Gillmor, C. S. in Current Topics in Developmental Biology Vol. 131 (ed. Grossniklaus, U.) 497–543 (Academic Press, 2019).
Hendelman, A. et al. Conserved pleiotropy of an ancient plant homeobox gene uncovered by cis-regulatory dissection. Cell 184, 1724–1739.e16 (2021).
Khanday, I. & Sundaresan, V. Plant zygote development: recent insights and applications to clonal seeds. Curr. Opin. Plant Biol. 59, 101993 (2021).
Khanday, I., Santos-Medellín, C. & Sundaresan, V. Somatic embryo initiation by rice BABY BOOM1 involves activation of zygote-expressed auxin biosynthesis genes. New Phytol. 238, 673–687 (2023).
Itoh, J.-I. et al. Genome-wide analysis of spatiotemporal gene expression patterns during early embryogenesis in rice. Development 143, 1217–1227 (2016).
Sato, Y. et al. A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl Acad. Sci. USA 93, 8117–8122 (1996).
Ito, Y., Eiguchi, M. & Kurata, N. KNOX homeobox genes are sufficient in maintaining cultured cells in an undifferentiated state in rice. Genesis 30, 231–238 (2001).
Shim, S. et al. Transcriptome comparison between pluripotent and non-pluripotent calli derived from mature rice seeds. Sci. Rep. 10, 21257 (2020).
Ueda, M. et al. Transcriptional integration of paternal and maternal factors in the Arabidopsis zygote. Genes Dev. 31, 617–627 (2017).
Eshed Williams, L. Genetics of shoot meristem and shoot regeneration. Annu. Rev. Genet. 55, 661–681 (2021).
Aflaki, F., Gutzat, R. & Mozgová, I. Chromatin during plant regeneration: opening towards root identity? Curr. Opin. Plant Biol. 69, 102265 (2022).
Kitagawa, M. & Jackson, D. Control of meristem size. Annu. Rev. Plant Biol. 70, 269–291 (2019).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Kim, J. B. et al. Oct4-induced pluripotency in adult neural stem cells. Cell 136, 411–419 (2009).
Radzisheuskaya, A. & Silva, J. C. R. Do all roads lead to Oct4? The emerging concepts of induced pluripotency. Trends Cell Biol. 24, 275–284 (2014).
Wei, X. et al. Synthetic apomixis with normal hybrid rice seed production. Mol. Plant 16, 489–492 (2023).
Li, D., Fan, L., Shu, Q. & Guo, F. Ectopic expression of OsWOX9A alters leaf anatomy and plant architecture in rice. Planta 260, 30 (2024).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497 (1962).
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA 74, 5463–5467 (1977).
Zhou, H., Liu, B., Weeks, D. P., Spalding, M. H. & Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 42, 10903–10914 (2014).
Rao, N. N., Prasad, K., Kumar, P. R. & Vijayraghavan, U. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc. Natl Acad. Sci. USA 105, 3646–3651 (2008).
Javelle, M., Marco, C. F. & Timmermans, M. In situ hybridization for the precise localization of transcripts in plants. J. Vis. Exp. https://doi.org/10.3791/3328 (2011).
Galbraith, D. W. et al. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049–1051 (1983).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Acknowledgements
We thank M. Talty and S. Tang for assistance with genotyping of rice plants, A. Gaikwad for help with plant growth and maintenance, and G. Austin for assistance with the rice transformations. This research was supported by grants from the National Science Foundation (no. IOS1936872) and the United States Department of Agriculture (USDA) Agricultural Experiment Station (AES) (project no. CA-D-XXX-6973-H) to V.S., and funds from the Department of Plant Sciences, UC Davis, and USDA AES (project nos CA-D-PLS-2736-H and CA-D-PLS-2812-RR) to I.K.
Author information
Authors and Affiliations
Contributions
V.S., I.K. and H.R. designed the study. I.K. and K.S. made the constructs for OsWOX9A ectopic expression. M.-J.C. and M.T. performed the rice transformations. H.R. performed all other experiments. H.R. analysed the data with V.S. and I.K. H.R., V.S. and I.K. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The University of California, Davis, has filed a patent application on efficient induction of parthenogenesis in crop plants (PCT/US2023/034142) arising from this work. The authors declare no other competing interests.
Peer review
Reviewer recognition
Nature Plants thanks Meng-Xiang Sun, Ok Ran Lee, Wei-Cai Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Phenotypic effects of pEC1.2::OsWOX9A expression and appearance of twin plants after haploid induction.
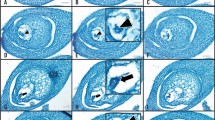
a. Expression of pEC1.2::OsWOX9A transgene resulted in smaller plants, whether in wild-type or in OsBBM1-ee background, suggestive of leaky vegetative expression from the pEC1.2 promoter. Left, OsWOX9A-ee + OsBBM1-ee diploid plant vs. OsBBM1-ee only diploid plant, OsWOX9A-ee + OsBBM1-ee plants are smaller compared to those with only the OsBBM1-ee transgene; right, OsWOX9A-ee + OsBBM1-ee diploid plant vs. OsWOX9A-ee only diploid plant, OsWOX9A-ee plant in OsBBM1-ee background has similar size compared to that in wild-type background. Scale bar, 5 cm. b. OsWOX9A-ee + OsBBM1-ee plants have narrower leaves compared to OsBBM1-ee only plants. c-d, The occurrence of twin progenies in parthenogenetic plants. Red arrowheads point to each single seedling. c. Twin plants at flowering stage. Left, haploid-haploid twin; right, haploid-diploid twin. d. Twin seedlings arising from a single seed.
Extended Data Fig. 2 Evolutionary relationship of WOX9 family proteins.
a. Alignment of a highly conserved 67 amino acid peptide domain of WOX9 family proteins, corresponding to amino acid residues 64 to 130 of rice OsWOX9A. This region corresponds to a subdomain of the homeobox originally identified in WUS, the first member of this protein family to be characterized. For reference, the alignment of OsWUS and OsWOX2 in this subdomain is shown in the bottom two rows. The red outlined box indicates the short peptide NVFYWFQN found in all the WUSCHEL-related proteins. However, the full 67 amino acid homeodomain is relatively poorly conserved in OsWUS, OsWOX2 and other WUSCHEL-related proteins families that are not WOX9 orthologs. b. A phylogenetic tree summarizing the evolutionary relationships between WOX9 family proteins constructed using PhyML3.0 (ref. 47). WOX9 gene duplications occurred independently in the cereal and Brassicaceae lineages.
Extended Data Fig. 3 Genotype of additional wox9awox9c mutant lines.
a. Sequence chromatograms at mutation sites and mutant DNA sequences of OsWOX9A and OsWOX9C in transgenic line WOX9AC#W3 compared to wild-type sequences. This line is biallelic for OsWOX9A with a 1 bp deletion null allele and a 6 bp in-frame deletion allele. For OsWOX9C, this line is biallelic with both alleles as 1 bp insertions. b. Sequence chromatograms and mutant DNA sequences at mutation sites in the OsWOX9A and OsWOX9C genes of transgenic line WOX9AC#501a, compared to wild-type sequences. Line WOX9AC#501a is biallelic for OsWOX9A with a 3 bp in-frame deletion allele and a 2 bp deletion null allele. For OsWOX9C, this line is biallelic with 10 bp and 26 bp deletion alleles. SgRNA protospacer sequences are shown in red boxes. The reverse primer was used for OsWOX9A sequencing results presented.
Extended Data Fig. 4 Histological analysis of T2 progeny embryos from T1 line WOX9AC#4-6, segregating for WOX9A but homozygous mutant for WOX9C.
a. Embryos from 3 DAP timepoint, before organogenesis stage. WOX9A/WOX9A wox9c/wox9c (left, n = 5/5) and WOX9A/wox9a wox9c/wox9c (middle, n = 6/6) embryos show normal development, while homozygous double mutant, wox9a/wox9a wox9c/wox9c (right, n = 6/6) embryos exhibit delayed development, showing smaller embryos with fewer cells. Scale bars, 100μm. b. Embryos from 5 DAP timepoint, after organ formation. WOX9A/WOX9A wox9c/wox9c (left, n = 4/4) and WOX9A/wox9a wox9c/wox9c (middle, n = 4/4) embryos show normal development, while wox9a/wox9a wox9c/wox9c (right, n = 2/2) embryos show arrested development. Co, coleoptile; ep, epiblast; lp, leaf primordia; ra, radicle; SAM, shoot apical meristem; sc, scutellum. Scale bars, 100μm. Arrested embryos appear to have no radicle, coleoptile, SAM, scutellum, or other structures. Instead, these embryos have only an epidermal layer, and the isotropic cells surrounding a core of more dense smaller cells inside. c. Embryos at 10 DAP, mature embryo developmental stage. WOX9A/wox9a wox9c/wox9c (left, n = 2/2) embryo shows normal development, while wox9a/wox9a wox9c/wox9c (right, n = 2/2) embryo shows arrested development. Scale bars, 400 μm.
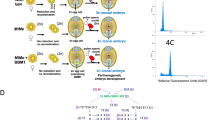
Extended Data Fig. 5 Co-expression of OsBBM1 and OsWOX9A using dexamethasone inducible glucocorticoid (GR) induction system.
a. Schematics of OsBBM1-GR and OsWOX9A-GR constructs. (b, c). Phenotype of seeds germinating on ½ MS media mock treated (b), or containing 10μM dexamethasone (c) (no other hormone added), for 2 weeks. From left to right, Wild-type (WT); OsWOX9A-GR; OsBBM1-GR; OsWOX9A-GR + OsBBM1-GR. Scale bars, 1 cm. Wild-type and OsWOX9A-GR germinated normally on dexamethasone (DEX) containing media, while OsBBM1-GR and OsWOX9A-GR + OsBBM1-GR formed calli from scutellum. Red arrowheads indicate shoots emerging from the seeds and white arrowheads point to calli. d. Expression of early embryo marker genes. RT-qPCR log2 fold changes in OSH1 and AMY1 transcripts is shown in OsBBM1-GR calli or OsWOX9A-GR + OsBBM1-GR calli treated for 2 weeks with 10μM dexamethasone. Embryogenic and non-embryogenic calli from scutellum of wild-type seeds were generated by callus induction media as previously described26. The log2 fold change is normalized to wild-type non-embryogenic calli. The height of columns represents mean values of the log2 fold changes, and error bars represent mean values ± SEM, calculated from three independent biological replicates and each data point represents the average of three technical replicates. Two-sided Tukey’s HSD test was used for the statistical analysis, and P values for each comparison are displayed in the graphs. Samples from left to right are: Wild type non-embryogenic calli; wild type embryogenic calli; OsBBM1-GR; OsWOX9A-GR + OsBBM1-GR.
Extended Data Fig. 6 Genotyping for pEC1.2::OsWOX9A and WOX9A WOX9C CRISPR-Cas9 segregating transgenic lines.
a. PCRs amplifying transgene-specific DNA fragment at the intersection of pEC1.2 promoter to OsWOX9A CDS with primers pEC1.2 Seq F and WOX9A Seq R were performed to genotype the parthenogenetic lines. RICE FLORICAULA LEAFY (RFL)43 (LOC_Os04g51000), a single copy gene, was used as the internal control. The fragment sizes of the amplicons - 539 bp for pEC1.2::OsWOX9A and 210 bp for RFL, respectively - were confirmed by comparison with the DNA marker. b. PCRs amplifying DNA fragment at HYGROMYCIN PHOSPHOTRANSFERASE (HPTII) gene were performed to confirm the presence of WOX9A WOX9C CRISPR construct, and RFL gene was used as an internal control. The fragment sizes of the amplicons - 321 bp for the CRISPR construct and 210 bp for RFL, respectively - were confirmed by comparison with the DNA marker. Primer sequences are provided in the Supplementary Table 6.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Tables 1–6.
Source data
Source Data Extended Data Fig. 6
Unprocessed gel.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ren, H., Shankle, K., Cho, MJ. et al. Synergistic induction of fertilization-independent embryogenesis in rice egg cells by paternal-genome-expressed transcription factors. Nat. Plants 10, 1892–1899 (2024). https://doi.org/10.1038/s41477-024-01848-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-024-01848-z
This article is cited by
-
Compound Inflorescence (S) represses fruit growth and seed development in tomato
Molecular Horticulture (2026)
-
Twin embryo formation by induced parthenogenesis
Plant Reproduction (2025)