Abstract
Plants display a remarkable regeneration capacity, which allows them to replace damaged or lost cells, tissues and organs, and thus recover from a broad spectrum of injuries1,2. Even lost stem cells can be regenerated from non-stem cells after competence acquisition, highlighting the enormous plasticity of plant cells. However, the molecular mechanisms underlying this process are still poorly understood. In the root, the highly conserved microRNA miR396 and its targets, the GROWTH-REGULATING FACTORs (GRFs), control the transition from stem cells to proliferative cells. miR396 promotes stem cell activity by repressing and excluding the GRFs from the stem cell area. In turn, the GRFs promote cell division in the proliferation zone3. Here we show that the miR396–GRF regulatory module guides stem cell reconstitution after root tip excision, playing a dual role: while miR396 promotes competence, the GRFs control regeneration speed. Moreover, plants with ectopic miR396 expression have defined stem cell niches before the excision but do not reconstitute them afterwards, remaining in an open state despite continuing to grow. We propose that this phenomenon is caused by dispersed stem cell activity, which supports growth after root tip excision without reconstituting the organized and spatially restricted stem cell niche typical of Arabidopsis roots.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The scRNA-seq data generated in this study are available at GEO under accession number GSE256274.
References
Ikeuchi, M., Ogawa, Y., Iwase, A. & Sugimoto, K. Plant regeneration: cellular origins and molecular mechanisms. Development 143, 1442–1451 (2016).
Ikeuchi, M. et al. Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 70, 377–406 (2019).
Rodríguez, R. E. et al. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell 27, 3354–3366 (2015).
Pérez-García, P. & Moreno-Risueño, M. A. Stem cells and plant regeneration. Dev. Biol. 442, 3–12 (2018).
Dolan, L. et al. Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84 (1993).
Campilho, A. et al. Time-lapse analysis of stem cell divisions in the Arabidopsis thaliana root meristem. Plant J. 48, 619–627 (2006).
Cruz-Ramírez, A. et al. A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol. 11, e1001724 (2013).
Rahni, R. & Birnbaum, K. D. Week-long imaging of cell divisions in the Arabidopsis root meristem. Plant Methods 15, 30 (2019).
Sena, G., Wang, X., Liu, H. Y., Hofhuis, H. & Birnbaum, K. D. Organ regeneration does not require a functional stem cell niche in plants. Nature 457, 1150–1153 (2009).
Efroni, I. et al. Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165, 1721–1733 (2016).
Zhou, W. et al. A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177, 942–956 (2019).
Matosevich, R. et al. Local auxin biosynthesis is required for root regeneration after wounding. Nat. Plants 6, 1020–1030 (2020).
Takahashi, N. & Umeda, M. Brassinosteroids are required for efficient root tip regeneration in Arabidopsis. Plant Biotechnol. 39, 73–78 (2022).
Heyman, J. et al. The heterodimeric transcription factor complex ERF115–PAT1 grants regeneration competence. Nat. Plants 2, 16165 (2016).
Durgaprasad, K. et al. Gradient expression of transcription factor imposes a boundary on organ regeneration potential in plants. Cell Rep. 29, 453–463 e453 (2019).
Rodríguez, R. E. et al. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112 (2010).
Horiguchi, G., Ferjani, A., Fujikura, U. & Tsukaya, H. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119, 37–42 (2006).
Kim, J. H., Choi, D. & Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104 (2003).
Liebsch, D. & Palatnik, J. F. MicroRNA miR396, GRF transcription factors and GIF co-regulators: a conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 53, 31–42 (2020).
Debernardi, J. M., Rodriguez, R. E., Mecchia, M. A. & Palatnik, J. F. Functional specialization of the plant miR396 regulatory network through distinct microRNA–target interactions. PLoS Genet. 8, e1002419 (2012).
Liu, D., Song, Y., Chen, Z. & Yu, D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 136, 223–236 (2009).
Galinha, C. et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057 (2007).
Aida, M. et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120 (2004).
Debernardi, J. M. et al. A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279 (2020).
Kong, J. et al. Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species. Front. Plant Sci. 11, 572319 (2020).
Luo, G. & Palmgren, M. GRF–GIF chimeras boost plant regeneration. Trends Plant Sci. 26, 201–204 (2021).
Bull, T. et al. GRF–GIF chimeric proteins enhance in vitro regeneration and Agrobacterium-mediated transformation efficiencies of lettuce (Lactuca spp.). Plant Cell Rep. 42, 629–643 (2023).
Feng, Q. et al. Highly efficient, genotype-independent transformation and gene editing in watermelon (Citrullus lanatus) using a chimeric ClGRF4–GIF1 gene. J. Integr. Plant Biol. 63, 2038–2042 (2021).
Yarra, R. & Krysan, P. J. GRF–GIF duo and GRF–GIF–BBM: novel transformation methodologies for enhancing regeneration efficiency of genome-edited recalcitrant crops. Planta 257, 60 (2023).
Chen, Z., Debernardi, J. M., Dubcovsky, J. & Gallavotti, A. Recent advances in crop transformation technologies. Nat. Plants 8, 1343–1351 (2022).
Hou, N. et al. Epigenetic regulation of miR396 expression by SWR1-C and the effect of miR396 on leaf growth and developmental phase transition in Arabidopsis. J. Exp. Bot. 70, 5217–5229 (2019).
Shahan, R., Nolan, T. M. & Benfey, P. N. Single-cell analysis of cell identity in the Arabidopsis root apical meristem: insights and opportunities. J. Exp. Bot. 72, 6679–6686 (2021).
Brady, S. M. et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007).
Han, E., Geng, Z., Qin, Y., Wang, Y. & Ma, S. Single-cell network analysis reveals gene expression programs for Arabidopsis root development and metabolism. Plant Commun. 5, 100978 (2024).
Heyman, J. et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342, 860–863 (2013).
Fulcher, N. & Sablowski, R. Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl Acad. Sci. USA 106, 20984–20988 (2009).
Desvoyes, B., Arana-Echarri, A., Barea, M. D. & Gutierrez, C. A comprehensive fluorescent sensor for spatiotemporal cell cycle analysis in Arabidopsis. Nat. Plants 6, 1330–1334 (2020).
Kim, J. H. & Lee, B. H. GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons and shoot apical meristem. J. Plant Biol. 49, 463–468 (2006).
Lee, S. J. et al. GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR specify meristematic cells of gynoecia and anthers. Plant Physiol. 176, 717–729 (2018).
Sabatini, M., Heidstra, R., Wildwater, M. & Scheres, B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17, 354–358 (2003).
Heidstra, R., Welch, D. & Scheres, B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18, 1964–1969 (2004).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Melsted, P. et al. Modular and efficient pre-processing of single-cell RNA-seq. Nat. Biotechnol. 39, 813–818 (2021).
Zhang, H. SCP: Single cell pipeline. R package version 0.5.6 https://github.com/zhanghao-njmu/SCP (2023).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies and species. Nat. Biotechnol. 36, 411–420 (2018).
Amezquita, R. et al. Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145 (2020).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Clough, S. J. & Bent, A. W. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis. Plant J. 16, 735–743 (1998).
Acknowledgements
We thank M. Bennet, C. Gutiérrez, B. Scheres, J. H. Kim and N. Han for providing seeds; S. Gornik for support with computational analyses; R. Vena for help with confocal image acquisition and analysis; D. Aguirre and K. Piiper for help with plant care; and members of our group for input on our work. J.L.B., F.E.L. and D.L. were supported by fellowships from CONICET, and J.F.P., R.E.R. and C.S. are members of the same institution. This research is supported by grants from Agencia I+D+i (Argentina, PICT-StartUp-2019-0000-2, PICT-2021-I-A-00513), CONICET (PUE 086), SF DTT-2023-072 and ICGEB (CRP/ARG17-01) and a Humboldt award to J.F.P., and the ERC synergy grant no. 810296 ‘DECODE’ to J.U.L.
Author information
Authors and Affiliations
Contributions
J.L.B. performed and analysed the experiments. J.L.B., F.E.L. and D.L. developed the plant materials. F.E.L., L.B., X.Z. and J.Z. performed the experiments. J.L.B., R.E.R., C.S., J.U.L. and J.F.P. conceived the experiments. J.L.B. and J.F.P. wrote the manuscript. R.E.R., D.L., C.S. and J.U.L. revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Reviewer recognition
Nature Plants thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Phenotype frequencies during regeneration in wild-type roots (complementary to Fig. 1).
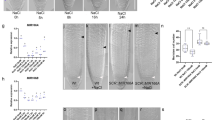
a Root meristem of WT Arabidopsis thaliana expressing the quiescent center marker pWOX5:GFP (magenta). Dotted white line outlines the stem cells. QC: quiescent center; SCN: stem cell niche. b Phenotype frequencies for WT after cutting at 100 µm from the tip. N indicates the number of experiments from which the frequencies were averaged; Total n indicates the total number of roots included in all said experiments. c Phenotype frequencies as in B, further classified into open or closed states. d Possible meristem states after cutting, according to cellular anatomy (middle and right panels are closeup of pictures in Fig. 1c). Dotted red and green lines follow the cortex cell files, that stay parallel in the open state but converge into isodiametric cells in the closed state. e Extreme cases of W50/W200 ratio: intact or fully regenerated roots are the most conical, while roots right after cutting are the most cylindrical.
Extended Data Fig. 2 GRF expression and activity during regeneration (complementary to Fig. 1).
a Regeneration of pGRF2:GRF2-GFP (magenta) line after cutting at 100 µm from the tip. b Ratio of width at 50 µm to width at 200 µm (W50/W200) for WT and p35S:GRF5 lines during regeneration. n represents the number of observed roots. Red asterisk indicates significant differences with WT (ANOVA followed by two-sided Tukey’s tests). c Regeneration of the same lines as in b after cutting at 100 µm. d Phenotype frequencies for the same lines as in b and c after cutting at 100 µm from the tip. White asterisk indicates significant differences with WT in closed-state proportion (χ2 test, α = 0.05, degrees of freedom = 1). Dashed red lines in a and c indicate cut position. White arrowheads mark the uncut or regenerated QCs. Error bars in b indicate standard error of the mean. WT data in b and d are the same as in Fig. 1b, d.
Extended Data Fig. 3 GRF gene expression levels in p35S:MIR396 lines (Complementary to Fig. 2).
a and b Relative expression of all nine GRF genes, measured by quantitative real-time PCR, in WT and both strong (a) and moderate (b) p35S:MIR396 lines. WT data are the same in both plots. Red asterisks indicate significant differences with WT (two-sided Kruskal-Wallis test). In each boxplot, the central line depicts the median of the data, the box covers the interquartile range, and the whiskers represent the maximum and minimum data points. n represents the number of samples taken.
Extended Data Fig. 4 Regeneration of a moderate p35:MIR396 line (complementary to Fig. 2).
a W50/W200 ratio in WT and a moderate p35S:MIR396 line during regeneration. n represents the number of observed roots. b Root elongation for control (uncut) and cut WT and moderate p35S:MIR396 lines after cutting at 100 µm from the tip. n represents the number of observed roots. Red asterisks indicate significant differences with uncut controls (two-sided repeated measures ANOVA, p < 0.01). c Regeneration of the same lines as in a and b after cutting at 100 µm from the tip. d WT and moderate p35S:MIR396 root tips during early regeneration after cutting at 100 µm, stained with Lugol’s solution to visualize starch granules. e Phenotype frequencies for WT, moderate and strong p35S:MIR396 lines after cutting at 100 µm from the tip. White asterisks indicate significant differences with WT in closed state proportion (χ2 test, α = 0.05, degrees of freedom = 1). N indicates the number of experiments from which the frequencies were averaged; Total n indicates the total number of roots included in all said experiments. f Frequencies for conical and cylindrical (W50/W200 ratio < 0.7 and ≥ 0.7, respectively) roots in the same lines as in e after cutting at 100 µm from the tip. White asterisks and n as in e. n represents the number of observed roots. Error bars in a, b and e indicate standard error of the mean. Dashed red lines indicate cut position. White arrowheads mark the uncut or regenerated QCs.
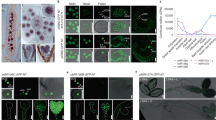
Extended Data Fig. 5 Ectopic mir396 expression increases number of cells with QC-associated gene expression (complementary to Fig. 3).
a UMAP of all root tip cells in our dataset, coloured by genotype and treatment. b Number of cells expressing QC-associated genes, as previously described33,34, in WT and p35S:MIR396 before (Control) and 5 days after cutting at 100 µm. Genes expressed in less than 50 cells were excluded. Black line indicates average of all included genes. c to g Boxplots (left) and UMAPs (right) split by treatment: WT uncut (Control) and 5 days after cutting (5DAC), and p35S:MIR396 Control and 5DAC, for the expression of selected QC-associated genes, namely: PLT1 (c), PLT2 (d), PLT3 (e), PLT4/BBM (f) and BRAVO (g). In each boxplot, the central line depicts the median of the data, the box covers the interquartile range, and the whiskers represent the maximum and minimum data points. n represents the number of cells expressing each gene (cells with NA value were filtered out).
Extended Data Fig. 6 WOX5 expression recovers earlier in a high-GRF line (complementary to Fig. 4).
pWOX5:GFP expression (magenta) in WT and pGRF3:rGRF3 during regeneration after cutting at 100 µm from the tip. Dashed red lines indicate cut position. White arrowheads mark the uncut QCs.
Extended Data Fig. 7 miR396 overexpression increases sensitivity to bleomycin and PLT2 expression pattern (complementary to Fig. 4).
a Length of zone showing cell death in WT and p35S:MIR396 roots after 24 h of mock or 2.4 mg/L bleomycin treatment. n represents the number of observed roots. Red asterisk indicates a significant difference with WT (right-sided Student’s t test). In each boxplot, the central line depicts the median of the data, the box covers the interquartile range and the whiskers represent the maximum and minimum data points. b pPLT2:CFP expression (magenta) in WT and p35S:MIR396 during regeneration after cutting at 100 µm from the tip. Dashed red lines indicate cut position. White arrowheads mark the uncut or regenerated QCs.
Extended Data Fig. 8 Regeneration stalling is not a mere consequence of meristem enlargement (complementary to Fig. 4).
a WT, p35S:MIR396 and pPLT2:PLT2-YFP (magenta) roots. b Meristem length in WT, p35S:MIR396 and pPLT2:PLT2-YFP lines. n represents the number of observed roots. Red asterisks indicate statistically significant differences with WT (two-sided ANOVA followed by Tukey’s comparisons, p < 0.05). c Regeneration of WT and pPLT2:PLT2-YFP roots cutting at 100 µm from the tip. Inset in the lower rightmost panel shows the QC with the YFP channel off. d Phenotype frequencies for WT and pPLT2:PLT2-YFP after cutting at 100 µm. n represents the number of observed roots. White arrowheads in a and c mark the uncut or regenerated QCs; black arrowheads mark the end of the meristem. Dashed red lines indicate cut position. In each boxplot in b, the central line depicts the median of the data, the box covers the interquartile range, and the whiskers represent the maximum and minimum data points.
Extended Data Fig. 9 High GRF activity shortens the regeneration competence zone (Complementary to Fig. 5).
a Regeneration of WT, mir396ab and pGRF3:rGRF3 during regeneration after cutting at 100 and 170 µm from the tip. b Phenotype frequencies for the same lines as in a after cutting at 100 and 170 µm from the tip. Black asterisks indicate significant differences with WT in the proportion of growing over total roots, and white ones, in the proportion of closed-state over growing roots (χ2 test, α = 0.05, degrees of freedom = 1). n represents the number of observed roots. Dashed red lines in a indicate cut position. White arrowheads mark the uncut or regenerated QCs. Images and data from cuts at 100 µm are the same as in Fig. 1d, e.
Supplementary information
Supplementary Information
Supplementary Table 1 legend.
Supplementary Table
Supplementary Table 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Baulies, J.L., Rodríguez, R.E., Lazzara, F.E. et al. MicroRNA control of stem cell reconstitution and growth in root regeneration. Nat. Plants 11, 531–542 (2025). https://doi.org/10.1038/s41477-025-01922-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-01922-0
This article is cited by
-
A microRNA defines root regeneration
Nature Plants (2025)
-
Root regeneration under miRNA control
Nature Plants (2025)
-
Genotype-dependent enhancement of tomato transformation and regeneration by GRF4 and GIF1
Plant Growth Regulation (2025)