Abstract
Replacing implanted medical hardware due to infection often requires one or more revision surgeries. Each surgery triggers a tissue injury response and disrupts the established bacterial biofilm. However, the complex tissue response to reinjury and biofilm disturbance is not well understood. Our results show that with an existing infection, immunological niches such as the bone marrow, lymph nodes, and circulating blood further upregulate pro-inflammatory programs in response to revision. Rather than reducing bacterial burden, this heightened inflammation provokes virulence factor expression and tissue damage, including bone osteolysis and muscle fibrosis. While muscle fibrosis appears transient and begins resolving by 14 days post-revision, osteolysis continues to progress. This study defines the timing and pathophysiology of coordinated multi-tissue responses to revision during infection. Understanding how host–pathogen interactions influence tissue recovery after revision can help identify risks and guide interventions that minimize damage and maximize bacterial clearance.
Similar content being viewed by others
Introduction
Surgery for traumatic fracture repair or arthroplasty often requires the implantation of stainless steel or titanium hardware components1. The use of implanted material in the surgical site drives an increased risk for infection as abiotic materials are rapidly coated with serum proteins, creating an ideal surface for bacteria to colonize and form a biofilm2. While free-floating (planktonic) bacteria are susceptible to immune defenses and antibiotics, bacteria in a biofilm community surrounded by a matrix of extracellular polymeric substances (EPS) are far less susceptible3,4,5,6,7. The EPS barrier is composed of proteins, polysaccharides, and nucleotides which limit the diffusion of antibiotics and physically block innate immune effectors from reaching the encased bacteria8,9. Bacteria within biofilms also employ active defenses by producing virulence factors which molecularly thwart the immune response and limit antibiotic efficacy, further promoting pathogen survival10,11,12,13. One of the most common biofilm-forming bacteria in implant-associated infection is Staphylococcus aureus (S. aureus), which includes both methicillin-sensitive (MSSA) and resistant (MRSA) strains2,14. In fact, antibiotic-resistant strains of pathogenic bacteria now constitute the majority of orthopedic infections, presenting significant clinical challenges that reduce the number of available treatment options15.
The standard treatment for infected hardware involves a revision surgery to remove dead and infected tissue, followed by replacement of the biofilm-coated hardware16,17. In the United States, a two-stage revision (2 surgeries) is the most common procedure16,17. At the first surgery, the infected implant is removed, and antibiotics are administered both locally (loaded within a cement spacer) and systemically to kill hidden bacteria18. Sometime later (usually several weeks), when the bacteria are considered cleared, a second surgery is performed to reimplant the new hardware18. In Europe, a one-stage revision surgery where the infected hardware is removed and replaced in a single procedure is more common16,17. A third option, called the “DAIR” procedure (debridement, antibiotics, and implant retention), where the implanted hardware remains in place, is also considered in certain cases19. The benefits of DAIR are a shorter surgical procedure, better preservation of the bone structure, and faster postoperative recovery. Unfortunately, this strategy results in comparatively lower success rates (30-80%)19. Conversely, both one-stage and two-stage revisions have broadly comparable patient outcomes with reinfection rates of 15-20%16,17,18,20. Importantly, one-stage revision is typically offered under stricter selection criteria (e.g., identified susceptible organism, good soft-tissue envelope, no sinus tract), while two-stage remains the historical ‘gold standard’ used for more complex or higher-risk presentations. This difference may explain why overall “success” can appear higher for two-stage despite similar reinfection figures in well-selected one-stage cohorts21,22. In all cases, patients are placed on long-term courses of systemic antibiotics combined with antibiotic-loaded cements to eliminate the remaining bacteria and prevent reinfection23.
While surgical intervention for infected hardware is the accepted standard of care, the process of making an incision and debriding necrotic tissue is not without consequences. The process of freshly wounding infected tissue not only activates local and systemic responses but also disrupts biofilms. Studies investigating the complex multi-tissue and bacterial responses to surgery – both immediately and over time – are limited. In fact, most studies only address the effects of infection and/or infection mitigation on bone-related processes. More recent studies on bone infection have detailed the ability of S. aureus to invade the osteocyte lacuna-canalicular network (OLCN) either directly through bone injury or indirectly through other mechanisms24,25,26. For example, infected “trojan horse macrophages” can carry bacteria into the marrow and release them25. With this access, individual bacteria can squeeze into and through the OLCN to infect osteocytes26. Osteocyte death then leaves empty lacunae which create pockets where dormant bacteria reside, becoming reactivated as bone is resorbed through remodeling24,25,26,27. These types of detailed studies have not been routinely extended to the surrounding periprosthetic tissues, nor do they explore the coordinated multi-tissue and systemic responses.
Indeed, the impact of implant and hardware-associated infection on other tissues beyond bone is not well understood. While it is known that infection and persistent inflammation can compromise bone marrow function, resulting in fibrosis and reduced hematopoietic function, the extent and timing of this disruption and how fast recovery occurs is unknown28. Interestingly, skeletal muscles, which lay in direct contact with bone and hardware-associated biofilms, are typically resistant to bacterial invasion29. However, during revision surgery, muscle fibers and fascial planes are separated or cut, providing access for bacteria to enter30. Despite this, how the muscle tissue and nearby immune cells respond to surgical injury and biofilm disruption remains less well known. Finally, while it is well established that S. aureus virulence factor expression is highly dynamic and can be dramatically affected by changes to the environment or biofilm disruption in vitro, little work has been done to investigate the in vivo response31,32. It is likely that the response to surgical intervention for infection is not determined by any one of these individual sites, but through the coordinated response of all affected tissues. Importantly, understanding each tissue’s response and contribution to infection eradication and healing could better predict the outcome of revision surgeries.
This investigation focuses on understanding the complex pathophysiology of host–pathogen responses to revision surgery across multiple tissue niches, both in direct contact with infected implants and at more distal sites. We hypothesize that surgical revision in the context of active infection provokes a coordinated, tissue-specific inflammatory response that fails to reduce bacterial burden but instead enhances bacterial virulence and drives long-term tissue damage. To test this, we used a rat model of surgical revision for hardware-associated biofilm infection and performed temporal profiling to capture the molecular interplay between S. aureus and surrounding tissues, including bone, muscle, bone marrow, lymphoid tissue, and blood. Taken together, the findings from this study may help inform the timing of interventions and strategies to prevent or mitigate some of the devastating outcomes following revision surgery.
Results
Bacteria colonize hardware and infiltrate all surrounding tissues within seven days of initial infection
To study the host immune and pathogen responses to infection in individual tissues after surgical revision, we developed a rat model of bone and hardware infection. In this model, at index surgery, infection is established by delivering 105 colony forming units (CFUs) S. aureus on a collagen sponge placed over an implant along the femur (Fig. 1A). The hardware, muscle, lymph nodes (draining and contralateral), and bone with marrow were assessed after 7 days to confirm biofilm growth and tissue pathophysiology (Fig. 1B). Established infection was grossly confirmed by the presence of necrotic tissue, swelling, and purulent material at the level of the muscle tissue or wound fluid at the level of the implants (Fig. 1C). Counts of 5-6 log10 CFU confirmed comparable bacterial colonization in both the muscle tissue and on hardware after 7 days of infection (Fig. 1D).
A Diagram of infection and end-term timeline. B Diagram of tissue collection protocol. C Representative gross images of uninfected and infected incision sites 1 week after index surgery. D CFU counts on the tissue and hardware 1 week after index surgery. E Representative image of infected muscle tissue 1 week after index surgery. Stained with Brown and Hopps. Visible S. aureus bacteria (dark purple) are marked with a yellow arrow. F Picrosirius red-stained muscle tissue (yellow) from uninfected and infected rats 1 week after index surgery highlighting myofascial fibrosis (red). G Quantification of depth of fibrosis (µm) in uninfected (n = 5) and infected (n = 7) rats determined from picrosirius-red-stained muscle sections. P-value determined using Mann Whitney test (p < 0.05). H Significantly upregulated gene ontology (GO) pathways from bulk RNA sequencing comparing infected (n = 5) to uninfected (n = 5) rats 1 week after index surgery. I Significantly downregulated GO pathways comparing infected to uninfected rats 1 week after index surgery. J Trichrome staining of the bone in uninfected rats with structural elements labeled: muscle (mu), center hole (h, outlined), reactive bone (rb), cortical bone (cb), bone marrow (ma), and growth plate (gp). Brown and Hopps staining of the bone with insets showing bone marrow with leukocytes and cortical bone with osteocytes. K Trichrome, Brown and Hopps staining of the bone in infected rats with labeled structural elements. Brown and Hopps insets show cortical bone with bacteria within osteocytes (yellow arrows), empty lacunae (*), and bone marrow with planktonic bacteria and bacteria engulfed by a phagocyte (yellow arrows).
We used muscle tissue histology to determine the extent of infection after 7 days. Sections stained with Brown and Hopps, a modified Gram stain, in Fig. 1E highlighted Gram-positive S. aureus bacteria (yellow arrows) within both fibrotic regions (found adjacent to the incision site) and deeper in the tissue between the muscle myofibrils. Picrosirius red staining delineated the red areas of muscle fibrosis (indicating tissue damage) from healthy muscle fibers (yellow) (Fig. 1F). Although some muscle tissue fibrosis is evident in uninfected rats after surgery alone, there is a significant ~3-fold increase in the depth of fibrotic tissue associated with infection (Fig. 1G)33,34,35. Molecular changes observed by RNA sequencing in the infected muscle compared to the uninfected after 7 days showed an upregulation of immune and inflammatory responses including: the adaptive immune response, innate immune functions like granulocyte migration, and the inflammatory response (Fig. 1H). Downregulated processes associated with infection primarily include muscle adaptation, vesicle transport, and energy homeostasis processes (Fig. 1I).
We next compared bone and bone marrow pathology after 7 days of infection using trichrome staining to assess structural changes and Brown and Hopps staining to detect S. aureus infiltration. In both infected and uninfected rats, the center hole (h, outlined) exhibits visible reactive bone (rb), confirming bone remodeling at the surgical implant sites (Fig. 1J, K). The cortical bone (cb) appears denser and less porous in the uninfected rats, whereas in the infected rats, the cortical bone is expanded and seems to exhibit more microcracks, which are stained dark purple with Brown and Hopps stain as the fuchsin dye lodges in the cracks and binds the exposed calcium-rich bone matrix (Fig. 1J, K)36. Compared to the bone marrow (ma) of healthy rats, which demonstrates uniform cellularity populated by viable cells, the marrow of infected rats shows depletion of cellular elements with increased fibrotic tissue (Fig. 1J, K). In infected rats, planktonic bacteria (yellow arrows) are present both dispersed throughout the bone marrow and engulfed within phagocytes (Fig. 1K). Bacteria are also observed within osteocytes and in empty lacunae (*) where healthy osteocytes usually reside (Fig. 1K). These data indicate that at 7 days post-infection, both bone and marrow are infiltrated with bacteria, with lesions indicating homeostatic disturbance and compromised tissue.
While inflammatory cytokines are likely to peak in the blood serum within 24–48 hours post-injury or infection, we used a cytokine array ELISA to determine if the systemic serum response persisted after 7 days (Table 1)37. Five cytokines were significantly upregulated after 7 days. Infected rats had significantly increased CINC-2 (~61 fold), ICAM-1 (~18-fold), and L-selectin (~4 fold). These cytokines typically upregulate neutrophil recruitment and inflammation and facilitate leukocyte recruitment from circulation to the site of infection38,39,40. IL-1α, a cytokine that triggers both local and systemic inflammatory immune responses, and TIMP-1, a growth factor that modulates MMP and immune cell activity, were also significantly upregulated 38- and 3-fold respectively41,42. Together, results from this model indicate that we have successfully established a robust infection comprising the hardware, bone, and surrounding tissues. Specifically, we observe early bacterial invasion of the bone and marrow, colonization of the OLCN, and loss of osteocytes that are characteristic of the pathology typically associated with a chronic orthopedic infection.
Invasion of osteocytes and marrow by S. aureus triggers progressive bone destruction and marrow fibrosis
After we determined that by 7 days a clear infection was established, we performed revision surgery. Adequate levels of infection were confirmed for each rat by determining the CFUs on the 2 screws implanted at index surgery. At each end-term timepoint (6 h, Day 4, and Day 14 post-revision), CFUs were measured from both the hardware placed at revision and the muscle tissue (Fig. 2A). There was a slight, but insignificant, decrease in CFUs at 6 h post-revision, which was possibly associated with the rinse and debridement procedures (Fig. 2B). By Day 4 the CFUs recovered to slightly above revision levels at ~6.5 log10 and remained at that level until Day 14 (Fig. 2B). These results suggest that our model generates a stable, non-resolving infection in both the muscle tissue and on the hardware.
A Diagram of experimental timeline with index surgery, revision surgery, and end term timepoints. B Average CFU measurements on the implant hardware and tissue over time in infected rats. Pre-revision n = 3, 6 h n = 6, Day 4 n = 4, Day 14 n = 6. Data are presented as mean ± SD. (C) Micro-CT scans of uninfected and infected femurs on Day 14 post-revision. D Quantification of cortical bone volume/total volume ratio in uninfected (n = 3) and infected (n = 3) rats on Day 14 post-revision. E SEM image of a biofilm on an implant on Day 14 post-revision. Inset shows S. aureus bound to an extracellular matrix (black arrow). White arrows mark structures consistent with EPS. Blue arrows mark neutrophils surrounding the biofilm. F Representative images of Brown and Hopps staining of femur mid-sections at all timepoints. Green arrows mark S. aureus. G Histological scoring of bacterial clumping and total infection score within the bone. For all timepoints n = 4. P-values determined using Kruskal-Wallis test (p < 0.05). H Representative TRAP-stained femurs showing osteoclasts in dark red. (I) Quantification of osteoclasts/mm2 of bone in uninfected (pre-revision n = 5, Day 4 n = 4, Day 14 n = 4) and infected (pre-revision n = 5, Day 4 n = 3, Day 14 n = 4) femurs. P-values determined using one-way ANOVA with multiple comparisons (p < 0.05).
An important clinical indication of biofilm infection is osteolysis43,44. To confirm this physiological endpoint in our model, we used micro-CT scanning to determine the extent of reactive bone formation and osteolysis at Day 14. There was a visible increase in both the reactive periosteal bone formation on the outside surface of the whole bone (shown in whole bone image) and bone loss by osteolysis (shown in the transverse section) in the infected femora compared to uninfected at Day 14 (Fig. 2C). Measurement of the bone volume to total volume (BV/TV) ratio confirmed a significant (p = 0.035) loss of cortical bone (Fig. 2D), but no significant change in reactive bone formation (data not shown). We further confirmed, using SEM to visualize the surface of the implanted lock-plate, the presence of a mature biofilm by Day 14 (Fig. 2E). S. aureus cocci (black arrows) are visible within a mesh-like extracellular material consistent with a biofilm matrix (white arrows), with neutrophils (blue arrows) at the periphery that do not appear to penetrate this layer (Fig. 2E).
We further determined bacterial localization using Brown and Hopps histological staining (modified Gram stain). On Day 4 post-revision numerous planktonic bacteria (green arrows) and some clumps are visible along the periosteum and within the OLCN (Fig. 2F). Larger clumps of biofilm were observed filling empty lacunae and coating the surfaces of dead bone fragments (Fig. 2F). The marrow on Day 14 was disrupted with large areas of fibrosis and numerous bacteria (Fig. 2F). Histological scoring (Supplementary Table 1) of bacterial presence in the bone (Supplementary Fig. 1) revealed a significant increase in bacterial clumping scores (p = 0.018) on Day 4, but no difference on Day 14 compared to pre-revision (Fig. 2G). The total infection score followed the same pattern with a significant increase from pre-revision to Day 4 (p = 0.045) and no significant difference between pre-revision and Day 14. These results suggest that bacterial colonization of the bone is a continuous process, but that a steady state may be reached over time. Finally, we used TRAP staining to visualize osteoclasts. These bone-resorbing cells are hyper-activated by bone injury and inflammatory cytokines45,46. As expected, in areas of bone damage due to screw insertion, or in areas of bacterial presence, large numbers of osteoclasts (dark red) are present regardless of the timepoint, with a significant increase (p < 0.001) observed on Day 14 (Fig. 2H, I). This likely accounts for the osteolysis observed and measured in bone at Day 14 post-revision (Fig. 2C, D). Taken together, these results show that both the bone and the bone marrow, a vital immunological niche, are compromised by bacteria, and show no signs of resolution.
Lymph nodes significantly expand after revision surgery
The compromised bone marrow niche due to infection led us to explore the draining lymph nodes – another immunological niche that is highly sensitive to both local injury and infection. To assess the differential effect of the revision surgery in the presence of S. aureus infection, we removed the draining popliteal lymph nodes from both the operated and non-operated limbs of the same rat and compared their weight at each timepoint (Fig. 3A, B). Pre-revision, the weight of the operated limb lymph node increased, but not significantly (p = 0.188) compared to the non-operated limb (Fig. 3B). At 6 h (p = 0.031) and Day 4 (p = 0.031) post-revision there were significant increases in the operated limb lymph node weights (Fig. 3B). While there was no significant difference on Day 14 between the operated and non-operated limb lymph nodes, there was greater variability in size and fewer retrievable samples in the non-operated limb (Fig. 3A, B). Surgery had no effect on lymph node weight in non-infected rat limbs (Supplementary Fig. 2).
A Gross images of draining popliteal lymph nodes from infected rats at each timepoint. B Measurement of draining popliteal lymph node weights in the infected and uninfected limbs of infected rats. Uninfected limb pre-revision n = 6, 6 h n = 6, Day 4 n = 6, Day 14 n = 3. Infected limb pre-revision n = 6, 6 h n = 6, Day 4 n = 6, Day 14 n = 4. P-values calculated using Kruskal-Wallis test (p < 0.05). C Diagram of rat lymph node structures used for orientation in the proximal histological analyses. D Brown and Hopps stained lymph nodes of infected rats at Day 4 and Day 14 showing the lymphatic vessels within the medulla and the capsule. S. aureus marked with yellow arrows. E H&E and Toluidine blue-stained lymph nodes in uninfected and infected rats at Day 4 and Day 14. H&E images show structure (nodule outlined, marked n, capsule marked c). Toluidine blue images are taken from the medulla (m) and show mast cells (dark purple). F Cells/µm2 in the lymph node medulla. N = 3 for all conditions. P-values calculated using Student’s t-test (p < 0.05). G Immunofluorescence of the medulla using antibodies against CD3 and CD4 (1:100). H Calculation of CD3+ and CD4+ cells/mm2 in the lymph node medulla. N = 3 for all conditions. P-values calculated using Student’s t-test (p < 0.05).
To further investigate the underlying cause of the surgically induced lymph node enlargement, we used multiple histological staining techniques to identify bacterial localization (Brown and Hopps), pathology (H&E), and mast cell presence (toluidine blue). Importantly, before beginning our analysis, an H&E-stained section of a normal rat lymph node was used to define the architecture, and each region was labeled (Fig. 3C). Representative Brown and Hopps sections show bacteria localized in the capsule and lymphatic vessels of the medulla on Day 4 (yellow arrows show planktonic bacteria) (Fig. 3D). By Day 14, bacteria are no longer detectable in the lymphatic vessels and are only visible in the capsular area (Fig. 3D). We then used H&E-stained histological sections to determine how the lymph nodes structurally and cellularly change in response to infection. Figure 3E shows representative high-magnification images of lymph nodes with the capsule (c), lymphofollicular nodules (outlines, n), and medulla (m) areas defined for infected and uninfected rats. Day 4 infected lymph nodes demonstrate features compatible with a drainage reaction including cortical follicular lymphoid hyperplasia with medullary plasmacytosis and expansion of the subcapsular and medullary sinuses by edema and cell infiltrates including plasma cells, Mott cells, macrophages, hemosiderophages, neutrophils, and mast cells (Fig. 3E). Toluidine blue staining showed an abundance of mast cells (dark purple) clustered together on both Day 4 and Day 14 (black arrows) in the medulla of infected lymph nodes, but dispersed mast cells present in uninfected lymph nodes (black arrows) (Fig. 3E). Finally, uninfected lymph nodes on both Day 4 and Day 14 had visible but less prominent lymphofollicular nodules with medullary sinuses containing scattered mast cells, macrophages, and few plasma cells (Fig. 3E). The infected lymph nodes on Day 14 have an intermediate pathology with reactive lymphofollicular nodules and plasmacytosis with mast cells, activated macrophages, and scattered neutrophils (Fig. 3E).
Because histological analysis revealed that infection led to congested sinuses, we used DAPI staining to determine the relative cellularity of the medulla in infected and uninfected lymph nodes. On Day 4, infected rats have significantly more (p = 0.031) cells/µm2 than uninfected rats, but by Day 14 there was no difference between the two groups (Fig. 3F). We next used antibodies against CD3 and CD4 to determine whether T cell population changes accounted for the change in lymph node size (Fig. 3G). On Day 4 there was a significant increase in both CD3+ (p = 0.008) and CD4+ (p = 0.032) T cell populations/µm2 in the medulla of infected rats compared to the uninfected (Fig. 3H). On Day 4 there are more CD4+ cells than CD3+ cells, which is unusual, but which is likely a population of CD3-CD4+ cells that are known to be active in lymph node development and repair47,48. By Day 14, there was only a significant increase (p = 0.038) in CD4 + T cells in the infected rats, and no difference in CD3 + T cells (Fig. 3H). This finding suggests that CD3+ and CD4 + T cell populations are at least partially responsible for the increased size of the lymph nodes by Day 4, but innate cells are not necessarily involved.
To determine systemic changes in the inflammatory landscape over time and explore how the infection-induced changes to the bone marrow and lymph node niches change inflammatory cytokine signaling, we measured the changes in circulating blood serum cytokines from revision to end term at Day 4 and Day 14 post-revision (Table 2). GM-CSF, which promotes development and activation of neutrophils and macrophages, is upregulated with infection at Day 4, but not at Day 1449. IL-2, which promotes T cell growth and regulates T cell differentiation is downregulated at Day 4, but not at Day 1450. The anti-inflammatory IL-10 and IL-13 cytokines are downregulated with infection at Day 14, but are not changed at Day 451,52. Pro-inflammatory TCK-1, which regulates macrophage and T cell activity and activates pro-inflammatory programs, is upregulated with infection at Day 14, but is not changed at Day 453. These findings suggest that there is still a circulating systemic inflammatory response at Day 14.
Revision surgery reactivates S. aureus proliferation and virulence factor expression, triggering an inflammatory response and muscle fiber loss
Revision surgery causes tissue injury and biofilm disruption. This makes the early events after revision surgery a critical period where muscle tissue mounts an inflammatory response and bacteria respond to physical disruption37. To assess these responses, we processed muscle tissue from the incision site for histology and both pro- and eukaryotic RNA isolation 6 h, 4 days, and 14 days post-revision. As established in Fig. 1F, a thick layer of muscle fibrosis is formed during the first 7 days of infection. We compared the pre-revision timepoint to the 6 h, Day 4, and Day 14 timepoints using picrosirius red staining. The incision site muscle (boxed, insets) at 6 h post-revision appears unchanged from the pre-revision sample (Fig. 4A, 1F). Of note, both samples have a clear, delineated line between the healthy yellow muscle fibers and the red areas of fibrosis35. This indicates that by revision or 6 h later, the muscle tissue fibrosis at the incision site is no longer infiltrative. However, on both Day 4 and Day 14 post-revision, the discrete layers of fibrosis (red) at the incision site (boxed insets) are transformed into a more infiltrative process that effaces the underlying degenerative muscle fibers (yellow) (Fig. 4A)35. In support of these observations, we measured the depth of the fibrotic region. We found that it significantly increases on Day 4 (p = 0.004) and Day 14 (p = 0.009) (Fig. 4B). This suggests that the process of revision reactivates muscle fibrosis.
A Representative image of picrosirius red-stained muscle tissue 6 h, Day 4, and Day 14 post-revision. B Quantification of depth of fibrosis (µm) in infected 6 h (n = 6), Day 4 (n = 6), Day 14 (n = 5) rats compared to pre-revision (n = 7) determined from picrosirius-red-stained muscle sections. P-values determined using one-way ANOVA (p < 0.05). C Significantly upregulated gene ontology (GO) pathways from bulk RNA sequencing comparing infected rats at 6 h (n = 5) to infected rats pre-revision (n = 5). D Significantly downregulated GO pathways comparing infected rats at 6 h to infected rats pre-revision. E Representative images of muscle stained with Brown and Hopps. Bacteria marked with yellow arrows. F Histological scoring of bacterial localization and bacterial clumping score within the muscle. Analysis compares 6 h (n = 6), Day 4 (n = 7), Day 14 (n = 8). P-values determined using the Kruskal-Wallis test (p < 0.05). G Relative virulence factor expression in vivo comparing pre-revision (n = 4), 6 h (n = 2), Day 4 (n = 5), and Day 14 (n = 5). P-values determined by ordinary one-way ANOVA (p < 0.05).
To determine the molecular events triggered in the muscle by the revision process, we used bulk RNA sequencing and the gene ontology (GO) library to compare differential expression between the infected rats 6 h post-revision and the infected pre-revision rats. Upregulated pathways 6 h post-revision include cellular responses to inflammatory IL-1, apoptotic signaling, and regulatory pathways for vasculature and cytokine production (Fig. 4C). Downregulated pathways include organization, structure, and regulation of actomyosin, cell migration, and mitochondria organization (Fig. 4D). These molecular changes correspond with the histological results that show revision surgery induces a renewed disruption of muscle fibers. Further, the molecular changes also reveal increased cell death, the inflammatory response, and angiogenesis in response to an incision.
To confirm the presence of bacteria in the region of renewed fibrotic activity, we used histology to describe bacterial localization/pathology within the tissue. Brown and Hopps staining showed the bacteria (yellow arrows) localized in both areas of active fibrosis and deeper in between muscle fibers (Fig. 4E). The muscle infection was scored based on key infection parameters (Fig. 4E, F) (Supplementary Fig. 3) (Supplementary Table 2). Bacteria were localized deeper within the muscle tissue at 6 h and only near the surface on Day 14 post-revision (Fig. 4F). Bacterial clumping, a measure of biofilm activity and infection severity, was highest on Day 4, but was again reduced by Day 14 (Fig. 4F).
When grown in vitro, biofilms have significantly higher expression of key virulence factors involved in quorum sensing and immune cell activation than planktonic bacteria (Supplementary Fig. 4). However, how S. aureus modulates the expression of these virulence factors in a complex in vivo environment has not yet been studied. Thus, we wanted to determine how the disruption of the S. aureus biofilm by revision would influence virulence factor expression. To track the changes in virulence factor expression in our model over time, we used a predefined, targeted RT-qPCR panel to determine the relative virulence factor expression at each time point (Fig. 4G). We did not perform whole-bacterium transcriptomics in tissue given the low pathogen RNS yield and host RNA dominance in vivo. Therefore, a focused virulence panel provided sensitive detection of early bacterial responses within this model. Superoxide Dismutase A (sodA) is a virulence factor responsible for detoxifying harmful reactive oxygen species (ROS) by splitting them into water and oxygen10. The agrB virulence factor is part of a quorum-sensing complex in S. aureus that allows the bacteria to sense and form biofilms11. SarA is a regulator of virulence factor transcription and controls expression of both sodA and the agr locus10. Both luk-PV and hla form pores in the membranes of attacking immune cells12,13. For each virulence factor, we saw the same pattern of expression: 6 h after revision surgery, the expression increased significantly from pre-revision levels (Fig. 4G). On Day 4 and Day 14 post-revision, virulence factor expression was significantly lower than at 6 h and not significantly different from pre-revision levels (Fig. 4G). Together, these findings suggest that revision surgery immediately triggers an enhanced inflammatory response and upregulated bacterial virulence 6 h post-revision. This translates to further destruction of muscle tissue fibers and the dissemination of bacteria back into the tissue which does not resolve by Day 14 post-revision.
Revision surgery reactivates the muscle inflammatory response
As the revision process has been shown to initiate muscle injury, angiogenesis, inflammation, and biofilm dispersal, we sought to investigate the specific interplay between the immune response and these events. RNA sequencing results from infected muscle 4- and 14-days post-revision were compared to the infected pre-revision muscle. On Day 4 there was upregulation of pro-inflammatory cell activity including leukocyte migration and proliferation, lymphocyte proliferation, and regulation of the immune effector process (Fig. 5A). We used RT-qPCR to confirm these results by assessing IL-1β and IL-6 levels and found that these were also upregulated on Day 4 in the infected rats (Supplementary Fig. 5). At the same time, metabolic pathways such as aerobic respiration, cellular respiration, and oxidative phosphorylation are downregulated (Fig. 5B). Pathways more consistent with muscle healing such as macromolecule biosynthesis and metabolic processing, not infection mitigation, are upregulated on Day 14, despite ongoing infection (Fig. 5C)54. However, indications of potential dysregulation in the healing outcome are seen in the downregulated Day 14 pathways which include negative regulation of apoptotic signaling, skeletal system development, and post-synapse assembly (Fig. 5D). All of these pathways are suggestive of muscle loss or destruction.
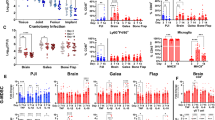
A Significantly upregulated gene ontology (GO) pathways from bulk RNA sequencing comparing infected rats at Day 4 (n = 6) to infected rats pre-revision (n = 5). B Significantly downregulated GO pathways comparing infected rats at Day 4 to infected rats pre-revision. C Significantly upregulated GO pathways comparing infected rats at Day 14 (n = 6) to infected rats pre-revision (n = 5). D Significantly downregulated GO pathways comparing infected rats at Day 14 to infected rats pre-revision. E GSVA analysis of bulk RNA sequencing over time for innate and adaptive cell signatures derived from PanglaoDB. F GSVA analysis of bulk RNA sequencing over time for MSigDB Hallmark Pathways. For all GSVA analyses, infected rats at pre-revision (n = 5), Day 4 (n = 6), and Day 14 (n = 6) are compared with uninfected rats at pre-revision (n = 5), Day 4 (n = 6), and Day 14 (n = 6) to generate a pattern of expression.
To more specifically query the immune cell dynamics at play over time in these large-scale pathway changes, we used GSVA analysis and the PanglaoDB55. This analysis indicates that both innate and adaptive immune cell populations are upregulated at each time point in the infected rats compared to the uninfected (Fig. 5E). However, the infected rats on Day 4 have the strongest and most coordinated upregulation of these immune cell populations. More specifically, GSVA using the hallmark pathways from the MSigDB56 indicate that the epithelial-mesenchymal transition, IL-6 JAK/STAT signaling, inflammatory response, and the interferon response are upregulated in infected animals, with the strongest and most coordinated response occurring again at Day 4 post-revision and becoming less coordinated between rats by Day 14 post-revision (Fig. 5F). These results suggest that there is an inflammatory immune response to infection and surgical revision, and that this response peaks 4 days after revision and starts to resolve by Day 14, despite the continued presence of bacteria in both the tissue and on the hardware.
To confirm cellular and hallmark pathway changes, we explored differences in protein levels of cytokines in the muscle between infected and uninfected rats at both Day 4 and Day 14 post-revision using a cytokine array Elisa (Table 3). Day 4 primarily shows upregulation of pro-inflammatory cytokines, whereas Day 14 primarily shows downregulation of pro-inflammatory cytokines and upregulation of anti-inflammatory cytokines (Table 3). Specifically, the pro-inflammatory cytokines PDGF-AA, Prolactin R, RAGE, and TCK-1 are downregulated at Day 14 compared to Day 4. Anti-inflammatory cytokines such as IL-4 are upregulated at Day 14 and not Day 4. These results further support the Day 4 peak in inflammatory activity following revision surgery that resolves by Day 14 in the muscle, despite ongoing infection.
Discussion
Here we investigate the response of multiple tissues to revision surgery in the presence of an S. aureus biofilm infection. Our study provides unique insights into the coordinated response of cells, organs, and S. aureus bacteria to surgical revision. We utilize a rat model to investigate the response of muscle, bone, bone marrow, draining lymph nodes, blood serum, and S. aureus to the combined insult of infection and surgical revision. The immediate (6 h) response is activation of S. aureus virulence factor expression and an inflammatory immune response in the surgically injured tissue that peaks ~4 days later. The presence of degenerative pathology occurs not only in the surgically injured muscle, but also in the bone, marrow, and lymph nodes. Apart from bone and marrow, degenerative and inflammatory pathology appears to be resolving by Day 14 post-revision in the soft tissues. Circulating blood cytokines confirm that surgical injury induces a systemic inflammatory response that begins resolving by Day 14. Unfortunately, this response fails to significantly reduce CFU counts across any time point. Instead, we observed a 6 h surge in virulence factor expression using a predefined, targeted RT-qPCR panel of virulence genes (e.g., soda, agrB, sarA, luk-PV, hla). Conclusions about bacterial behavior are accordingly bound to this panel, while host responses are informed by bulk RNA-seq. Despite this limitation, this work highlights the importance of employing productive strategies at the time of revision to target the reactivated bacteria and aid immune cells in a tissue-specific manner to promote more effective bactericidal strategies and protect tissue from injury.
In this study, we model a peri-implant infection in rats to understand the interaction between cells, tissues, and bacteria after surgical intervention. A successfully established infection is present after one week with gross clinical signs including pus, redness, and seeping wound fluid. Further, both the tissue and hardware at the incision site show CFU counts, which confirm a robust infection. It is important to note that we have not provided any antibiotic or alternative treatments for infection to directly assess tissue-pathogen responses. To this end, the rats mount a typical immune response consisting of upregulated adaptive and innate immune signatures, degenerative muscle fibrosis, and increased bone pathology, which all mimic patient pathology43,44.
Uninfected rats have healthy bone and marrow niches with inactivated leukocytes and healthy osteocytes. Infected rats, however, have visible S. aureus infiltration within the bone and marrow spaces one week post-infection. This finding coordinates with the bone infection pathology in other studies, but occurs earlier than has been previously described57,58,59. Typically, reports show that S. aureus infiltrates osteocyte lacunae and triggers osteocyte death, leaving holes in the bone after 14–28 days22,57,58,59,60,61,62. We see this occurring in areas of bone damage after only 7 days of infection, and the pathology continues to progress up to 14 days post-revision. By Day 14 S. aureus completely infiltrates the bone and marrow, leading to severe osteolysis and bone marrow fibrosis. This pathology closely resembles that seen in clinical cases of implant-associated infection43,44. The marked tissue damage we observe highlights that each additional surgical insult could compound inflammatory injury, explaining some of the limited clinical gains seen with two-stage revision when compared to one-stage revision.
Lymph nodes represent an intersection between bacteria and the immune response. In our model, only the combination of infection and revision surgery causes the lymph nodes to expand in size. Uninfected lymph nodes do not expand in response to surgical revision. The expansion of the lymph nodes is sustained from 6 h to Day 4, but resolves by Day 14 post-revision. The expansion of infected lymph nodes in response to revision is due to increased cellularity in part caused by the expansion of CD4 + T cells. Interestingly, CD4 + T cell populations are typically at their peak in the lymph node one week post-infection, but in our model, we see high numbers of CD4 + T cells at both 11 (Day 4) and 21 days (Day 14) after the initial infection63. This suggests that revision surgery in infected rats triggers CD4 + T cell expansion. Large numbers of CD4 + T cells can cause congestion in the lymph node, and previous studies show that this congestion can hinder cytotoxic CD8 + T cell activation and thus can reduce the efficacy of the immune response64. It has also been shown that lymph nodes that are expanded in size can have reduced lymph flow, contributing to slower and less effective bacterial clearance65. This supports the idea that revision surgeries have negative repercussions that prevent productive bacterial clearance, and thus reducing the number of revision surgeries may be beneficial to healing.
Interestingly, 6 h after revision surgery, bacterial CFUs are slightly reduced, and immune activation is increased. This is accompanied by a significant increase in bacterial virulence factor expression as bacteria are disrupted from the biofilm state, where low levels of virulence factors are secreted, to a metabolically active state that induces higher levels7,66,67,68,69. This transient increase in virulence rapidly drops by Day 4 post-revision. This finding suggests that the bacteria reenter a quiescent state after disruption by revision surgery. The progression of bacteria through these stages has several implications. The first is that surgical revision increases the potential for bacterial virulence, which has broad-reaching effects, but typically causes damage to immune cells and tissue66. The second implication is that revision surgery does appear to open a window of opportunity for effective treatment of the bacteria. In a quiescent/biofilm state, bacteria are less susceptible to both antibiotics and immune cell infiltration9,70. When disturbed and more metabolically active, the bacteria are more susceptible to treatments71. These findings suggest that revision may create a narrow window of enhanced susceptibility when S. aureus transitions out of its biofilm state and expresses virulence factors, during which a targeted antimicrobial intervention may be more effective.
The injury of infected muscle tissue in our model also indicates a more robust immune response after revision. On Day 4 post-revision, we find that the inflammatory response peaks and progresses toward resolution by Day 14 in the presence of bacteria. This increase in immune cell infiltration is accompanied by increases in muscle fibrosis on Day 4 and Day 14 when compared to uninfected controls. This is accompanied by an increase in the depth of infiltrating bacteria over time. These results correspond with previously published data which show that immune responses to infiltrating bacteria in skin and muscle are often self-limiting regardless of whether they successfully clear the bacteria72,73. Skeletal muscle tissue in particular is capable of sustaining low, but constant levels of inflammation in the presence of a persistent biofilm, while still initiating healing programs74. The immune response resolution that we see in our model could be due to the bacteria returning to its non-inflammatory biofilm state, the natural self-limiting course of the inflammatory response, or a combination of both. While we are unable to determine the exact cause of the immune response resolution, we can conclude that revision surgery is an inflammatory event that triggers upregulated inflammation beyond that which is present with the infection alone. This finding further supports that each revision comes with negative repercussions, and reducing the number of revision surgeries can prevent repeated inflammatory events that cause tissue damage. Additionally, we are able to conclude that a single revision surgery does contribute to an improvement in muscle pathology over time, and therefore, a single robust inflammatory event may be helpful in reducing the bacteria and improving muscle pathology.
Our study does have limitations. One limitation is the small sample size. Due to the challenging nature of the surgeries and that the rats used in this study are not genetic clones, there is increased variability in our results. While this variability does more closely mimic the range of clinical findings due to genetic heterogeneity, this also makes it challenging to draw broad conclusions based on these results. Another limitation of this study is that we do not have a time point within the first week of infection. Looking at the initial immune response to the index surgery and infection would provide insights into whether we are re-exacerbating an earlier immune response with the revision surgery, or whether we are generating a unique immune response with revision. We also, based on the nature of tissue sampling, are unaware of whether other parts of the host body are compensating for the defects in the infected limb. While we see no differences in the lymph nodes of the contralateral uninfected limb, we do not know whether the bone, marrow, muscle, or circulating immune system are able to compensate for the defects in the injured and infected limb long enough for that limb to recover.
Despite these limitations, our study contributes to the body of work that suggests one-stage revision is a viable and perhaps preferable, option for resolving implant-associated infections over the traditional two-stage revision. We show that a single revision surgery triggers an upregulation in bacterial virulence and the inflammatory immune response, which both highlight the short window of opportunity to administer effective treatment and lead to damage in the host tissues. By exploiting this knowledge, we suggest that clinicians may be better able to tailor their approach to one-stage revisions to include an aggressive, multi-faceted antibacterial regime at the time of revision to target the bacteria when they are most disturbed and susceptible to treatment. An enhanced, but rapidly resolving, immune response may benefit this system as it would be able to infiltrate the biofilm when it is disturbed 6 h after revision. Together, our data provide insights into the communication between tissues and bacteria during a hardware-associated biofilm infection and informs on the most effective timing and revision type for improving patient outcomes.
Methods
Inoculum Preparation
Frozen stock solution of Staphylococcus aureus (ATCC 25923) of a known concentration was thawed, and 50 µL was added to 10 mL sterile room-temperature Lactated Ringers solution (Medex Medical Supply). This volume has previously been shown to result in a concentration of 100,000 cells/250 µL solution. The suspension was vortexed, and five serial dilutions were created and plated on blood agar. These plates were placed in an incubator at 37 °C, and the growth was quantified the following day.
Orthopedic Animal Model
All procedures were approved by the University of Pennsylvania IACUC protocol #806810. The animals used in this study were 10-11-week-old male Sprague-Dawley rats. For index surgery, the animals were anesthetized via cage induction using Isofluorane in oxygen, and preoperative medications included meloxicam (Loxicom, 2 mg/kg SC), buprenorphine extended-release injectable suspension (Fidelis Animal Health Inc., 0.65 mg/kg SC), and Lactated Ringers solution (Medex Medical Supply, 5 mL SC). The left hind limb was circumferentially clipped and aseptically prepared. An incision through the skin was made from the level of the femorotibial joint to the coxofemoral joint following the path of the femur. The lateral surface of the femur was exposed, using a combination of blunt and sharp dissection to divide the vastus lateralis and the biceps femoris. Single bicortical 0.8 mm stainless steel screws (RISystem AG, Switzerland) were placed at the proximal and distal ends of the exposed femur. A unicortical hole was created in the mid-diaphyseal region and left unfilled to establish medullary access. A collagen sponge (Davol Inc., Becton, Dickinson and Company) was placed over the screw heads and empty hole and saturated with 250 µL of saline containing 105 CFU of S. aureus ATCC 25923. The muscle was closed in a simple continuous pattern using 4/0 polyglactin 910 (PDS, Ethicon, JnJ MedTech). The skin was closed in a simple continuous pattern using 4/0 polyglecaprone. Prior to skin closure the peri-incisional area was infiltrated with bupivacaine liposome injectable suspension (Nocita, Elanco).
Revision surgery was performed seven days after the index surgery. The animals were anesthetized and medicated as described above. An identical approach to the lateral femur was made and the screws were removed and used to calculate CFU counts at revision. The infected tissue was gently debrided. The incision was lavaged with 0.3% iodine solution with a contact time of 5 minutes. An 8-hole polyetheretherketone (PEEK) RatFix plate (RISystems AG, Switzerland) was placed on the lateral aspect of the femur and secured with 4 screws that avoided the locations of the previously placed screws. Closure of the soft tissues and local analgesia were performed as described above.
The animals were euthanized via intracardiac injection of a pentobarbital-based euthanasia solution (Vortech Pharmaceuticals Ltd.) and bilateral pneumothorax following anesthesia via cage induction using Isofluorane and oxygen either 6 hours (6 h), 4 days, or 14 days after revision surgery. All implants, the musculature cranial and caudal to the femur, the popliteal lymph nodes, and the femur were collected aseptically after confirmation of death.
Scanning Electron Microscopy
The hardware (screws and plate) was removed from the bone and fixed in 4% paraformaldehyde for at least 24 h. After fixation, the hardware was serially dehydrated using a series of graded alcohols from 70% to 100% with each phase lasting at least 6 h. After dehydration, the hardware was dried for 72 h. The samples were sputter-coated with gold and imaged using a Tabletop Hitachi Scanning Electron Microscope.
Bacterial culture and CFU calculations
Bacterial culture
An overnight culture was generated by taking one colony of S. aureus (ATCC 25923) from a streaked agar plate, adding it to 5 mL of sterile TSB, and incubating with shaking 12-18 hours overnight at 37 °C. A subculture was prepared by adding 1 mL of the overnight culture into 4 mL of sterile TSB and incubating with shaking at 37 °C for 2–5 hours. A densitometer was used to prepare a 108 CFU/mL solution, which was then diluted using sterile TSB to a 105 CFU/mL solution. 1 mL of the 105 CFU/mL solution was added to the well of a 24-well plate (Fisher Scientific) containing a 3D printed 1.5 cm diameter polycaprolactone (PCL) disc. The plate was incubated at 37 °C for 48 hours to generate biofilms on the PCL discs. In Vitro CFU Calculations: After 48 hours of incubation, the discs containing the biofilms were rinsed in sterile PBS (Thermo Fisher) and placed into a new 24-well plate containing 1 mL of sterile PBS. The discs containing the biofilms were sealed securely within the wells of the plate using parafilm (Sigma-Aldrich) and sonicated using the Branson CPX2800 ultrasonic bath for 20 minutes. The samples were diluted 1:100 by pipetting 20 μL of the sample into 180 μL of sterile PBS. The samples were serially diluted 7 additional times for a total of 8 different concentrations. Each sample was serially diluted in triplicate. 100 μL of each dilution was plated onto a Petrifilm (Fisher Scientific). The Petrifilms were incubated at 37 °C for 18-24 hours. CFU/mL counts were calculated based on manufacturer instructions. In Vivo CFU Calculations: The recovered implants were placed in sterile tubes and washed three times with sterile dPBS. The implants were placed in 3% Tween 20 (Fisher Scientific) and bath sonicated at 37 °C for 10 minutes. Serial dilutions of these fluids were plated on Petrifilms (Fisher Scientific), which were incubated 18–24 hours at 37 °C. The muscle tissue was vortexed in 5 mL sterile PBS for 5 minutes, and this fluid was handled identically to the implant fluid. CFU/mL counts were calculated based on manufacturer instructions.
RNA Isolation
Prokaryotic RNA isolation
Biofilms grown on discs were removed from TSB and washed using 2 mL PBS. 3 discs were combined in a 50 mL conical tube with 2 mL of Trypsin (Thermo Fisher), and discs were incubated with shaking at 37 °C for 20 minutes. Trypsin-containing bacteria were centrifuged for 4 minutes at 10,000 x g. The pellet was resuspended in 500 μL TE buffer (Fisher Scientific). Planktonic bacteria from a subculture were centrifuged for 4 minutes at 10,000xg. The pellet was resuspended in 500 μL TE buffer. Samples in TE buffer were centrifuged for 3 minutes at 8000xg at 4 °C. 100 μL of RNAse-free water was added to the pellet, and the pellet was vortexed vigorously for 3 minutes. 100 uL of TRIzol reagent (Fisher Scientific) and 100 μL of chloroform were added to the pellet and vortexed for 1 minute. The resulting mixture was incubated at 70 °C for 30 minutes. The samples were centrifuged at 12,000 x g for 10 minutes at 4 °C. 100 μL of the resulting aqueous solution was transferred to a new tube, and 200 μL of isopropanol was used to precipitate the RNA. After vigorous vortexing, the samples were centrifuged at 12,000 x g for 10 minutes at 4 °C. The isopropanol was removed, and 200 μL of 70% ethanol was added to the pellet. The pellet was vortexed and centrifuged for 5 minutes at 8000xg. The ethanol was removed, and the samples were dried for 5 minutes. RNA was resuspended in 20 μL of RNAse-free water. Eukaryotic RNA Isolation: Incision site tissue from the biceps femoris was excised, flash frozen in liquid nitrogen, and crushed. The muscle tissue was suspended in TRIzol reagent (Invitrogen) at a ratio of 1 mL reagent per 0.1 g tissue. Tissue was dissociated using M tubes and a gentleMACS tissue dissociator (Miltenyi Biotec). Chloroform phase separation was used to extract the RNA-containing aqueous layer and exclude protein from the dissociated tissue. A Macherey-Nagel Nucleospin RNA Midi Kit (Macherey-Nagel) was used to isolate the RNA.
RT-qPCR
Total RNA was converted to cDNA using RNA to cDNA EcoDry Premix (Takara Bio). 2 µL of cDNA was used for each RT-qPCR reaction. Reactions were performed in triplicate for each animal sample using SYBR Green Master mix and the QuantStudio 3 Real-Time PCR machine (Thermo Fisher) following the manufacturer’s instructions. Relative gene expression for each sample (primer details in Supplementary Table 3) was determined by normalizing to the expression of endogenous β-actin in each sample. Log2Fold Change was determined by comparing gene expression in the operated ipsilateral limb to the expression in the non-operated contralateral limbs of uninfected rats. Relative expression was determined using the 2-ΔΔCt method75.
RNA sequencing and analysis
RNA samples were sent to an off-site laboratory (Novogene) for quality validation and sequencing. The sequencing data were converted to raw read counts and mapped to the Rattus norvegicus genome by Novogene. Differential gene expression, gene ontology, and robust principal component analysis were completed using R Statistical Software version 4.2.2 using the DESeq2, clusterProfiler, and rrcov packages76,77,78. The transcripts of approximately 32,500 genes were measured, normalized, and the variance was visualized using robust principal components analysis. The identified outliers were discarded, and the remaining samples were analyzed to determine differentially expressed genes (DEGs). The significant DEGs presented an adjusted p-value < 0.05 using the Benjamini-Hochberg adjusted value. Significantly up- and downregulated genes were graphed using clusterProfiler, org.Rn.eg.db, ggplot2, patchwork, and scales libraries77,79,80,81,82. Gene set enrichment analysis (GSEA) and gene set variance analysis (GSVA) were conducted and graphed using the biomaRt, fgsea, GSVA, paletteer, and pheatmap libraries83,84,85,86,87. All data used in this study can be accessed through the GEO accession number: GSE301856.
Histology
Tissue preparation
Soft tissue and bone were excised and fixed immediately in 4% paraformaldehyde (PFA) for up to 48 h and transferred to 70% ethanol for 24 h. Bone was then placed in 14% ethylenediaminetetraacetic acid (EDTA) for 3 weeks for demineralization. All tissues were progressively dehydrated in graded alcohols and xylenes before being embedded in paraffin wax blocks (Thermo Fisher). Picrosirius Red: Picrosirius red stain (Thermo Fisher) was used to visualize and differentiate fibrotic tissue (red) and muscle tissue (yellow). The incision site fibrotic region was traced in its entirety, and the average depth of the fibrotic tissue, or thickness, was determined for each individual rat. Fat area per total fibrotic area was calculated by using 5 non-overlapping 20x images of the incision site per rat and determining the total fat area (white) per total fibrotic area (red), and generating the average %fat per animal. Myofibrils were calculated by using 5 non-overlapping 20x images of the incision site per rat and determining the total myofibril area (yellow) per total fibrotic area (red,) and generating the average %myofibrils per animal. All images were taken using a Nikon Eclipse E800 microscope. Color analysis was conducted using Image-Pro Plus 7.0 software. Brown and Hopps: Brown and Hopps staining kit (Newcomer Supply) was used to visualize the Gram (+) S. aureus (dark purple) within different tissues (muscle and bone). Infection parameters and total infection score for the muscle were determined using the scoring criteria outlined in Supplementary Table 2. Infection parameters and total infection score for the bone were determined using the scoring criteria outlined in Supplementary Table 1. All images were taken using a Nikon Eclipse E800 microscope. Color analysis was conducted using Image-Pro Plus 7.0 software. Hematoxylin and Eosin (H&E): H&E staining and pathology were conducted according to the University of Pennsylvania New Bolton Center histology core protocols. Lymph node slides were imaged at 10x to capture the structure of the capsule, nodules, and medullary space. The pathology report was created using a survey of 40x images. Toluidine Blue: Toluidine blue staining (Toluidine Blue O, Thermo Fisher, 1.1 mg/mL in 1% NaCl, 8% ethanol) was used to visualize mast cells (dark purple) within the lymph node. The medullary area was imaged at 20x. TRAP: Distal femur sections were deparaffinized and rehydrated. Sections were stained using Leukocyte Acid Phosphatase (TRAP) Kit (Sigma-Aldrich). We diverged from the kit protocol in the last stages of staining and counterstained with Fast Green FCF powder (Sigma Aldrich) instead of the kit hematoxylin solution. To count osteoclasts, stained sections were manually scanned under a Nikon Eclipse E800 microscope using a 40x objective lens. Dark red-stained osteoclasts in direct contact with the endosteum of the diaphysis were counted and presented as the number of osteoclasts per millimeter of bone perimeter. Bone perimeter was calculated using a spatial calibration function in Image-Pro Plus 7.0 software. All custom macros for Image-Pro Plus 7.0 software are available upon responsible request from the corresponding author.
Immunofluorescence
Muscle and lymph node sections were deparaffinized, rehydrated, and stained at the University of Pennsylvania Center for Musculoskeletal Disorders Histology Core (PCMD Admin Affiliate Member Award grant number: P30AR069619). Primary antibodies included 1:100 mouse anti-rat CD3 (Thermo Fisher) and 1:100 FITC-conjugated mouse anti-rat CD4 (Thermo Fisher). The secondary antibody used was Alexa Fluor 546 (Thermo Fisher).
Protein extraction
Protein was isolated from muscle tissue using tissue protein extraction reagent (TPER) (Thermo Fisher) and M tubes (Miltenyi Biotec) to mechanically homogenize the tissue using a gentleMACS tissue dissociator (Miltenyi Biotec). The homogenate was centrifuged at 1000 x g for 5 minutes, the supernatant was collected, and the supernatant was centrifuged again at 10,000 x g for another 5 minutes. The supernatant was collected and protein concentration was determined using the BCA assay (Thermo Fisher).
Cytokine array
Rat blood serum samples and isolated muscle proteins (n = 3 for each treatment group and timepoint) were sent to an off-site laboratory (RayBiotech), and quantitative proteomics were performed using a Miltiplex ELISA panel of rat-specific cytokines. Expression data for each treatment group (uninfected and infected) and time point (Day 4 and Day 14) were averaged, and a Student’s t-test was used to determine significant differences in expression.
Micro-CT scanning
Rat femurs were scanned using a Scanco Medical microCT 40 machine. Scans were performed along the long axis of the diaphysis using an energy of 55 kVp, a current of 145 mA, and a 200-ms integration time to produce a resolution of 16 mm3 voxel size. 140 slices were used to evaluate each hol,e with 20 additional slices on either side. All microCT data is reported as a percentage of bone volume at these holes. The manufacturer's software was used to calculate bone volume and mean density (BV/TV) data. The control femurs that did not undergo surgery were measured in their entirety for use as a reference for normalization. This negated minor variations in hole placement from sample to sample. Proximal and distal holes were grouped separately due to substantial differences in healing between locations.
Statistics
Graphs and statistical analysis were performed using R statistical software version 4.2.2 and the ggplot2, forcats, stringr, patchwork, scales, paletteer, and pheatmap libraries, or GraphPad Prism 10.4.180,81,82,86,87,88,89. Significant difference was defined in all cases as p < 0.05. The Mann-Whitney test was used in the analysis for BV/TV calculations in bone, CFU calculations in vivo, and RT-qPCR analysis. Student’s t-test was used for cytokine analysis, and lymph node immunofluorescence analysis. Kruskal-Wallis with False Discovery Rate was used for bone and muscle histology scoring analysis and lymph node weight analysis. An ordinary one-way ANOVA was used for virulence factor analysis, depth of fibrosis analysis, and osteoclast analysis.
Data availability
All data is freely available on reasonable request from the corresponding author. All sequencing data used in this study can be accessed through the GEO accession number: GSE301856.
Code availability
The underlying code for this study is not publicly available but may be made available to qualified researchers on reasonable request from the corresponding author.
References
Parry, M. C. & Duncan, C. P. The challenge of methicillin resistant staphylococcal infection after total hip replacement OVERLOOKED OR OVERSTATED?. Bone Jt. J. 96, 60–65 (2014).
Gupta, T. T. et al. Staphylococcus aureus aggregates on orthopedic materials under varying levels of shear stress. Appl. Environ. Microbiol. 86, e01234–20 (2020).
Costerton, J. W., Stewart, P. S. & Greenberg, E. P. Bacterial biofilms: a common cause of persistent infections. Science (1979) 284, 1318–1322 (1999).
Prosser, B. L., Taylor, D., Dix, B. A. & Cleeland, R. Method of evaluating effects of antibiotics on bacterial biofilm. Antimicrob. Agents Chemother. 31, 1502–1506 (1987).
Rabin, N. et al. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 7, 493–512 (2015).
Cangui-Panchi, S. P. et al. Battle royale: Immune response on biofilms – host-pathogen interactions. Curr. Res. Immunol. 4, 100057 (2023).
Dastgheyb, S. S. & Otto, M. Staphylococcal adaptation to diverse physiologic niches: an overview of transcriptomic and phenotypic changes in different biological environments. Future Microbiol. 10, 1981–1995 (2015).
Mah, T.-F. C. & O’toole, G. A. Mechanisms of Biofilm Resistance to Antimicrobial Agents. TRENDS in Microbiology vol. 9 http://tim.trends.com0966 (2001).
Di Martino, P. Extracellular polymeric substances, a key element in understanding biofilm phenotype. AIMS Microbiol. 4, 274–288 (2018).
Ballal, A. & Manna, A. C. Regulation of Superoxide Dismutase Genes by SarA in Staphylococcus aureus. J. Bacteriol. 191, 3301–3310 (2009).
Ji, G., Beavis, R. & Novick, R. P. Bacterial interference caused by autoinducing peptide variants. Science (1979) 276, 2027–2030 (1997).
Alonzo, F. & Torres, V. J. The bicomponent pore-forming Leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 78, 199–230 (2014).
Bartlett, A. H., Foster, T. J., Hayashida, A. & Park, P. W. α-Toxin Facilitates the Generation of CXC Chemokine Gradients and Stimulates Neutrophil Homing in Staphylococcus aureus Pneumonia. J. Infect. Dis. 198, 1529–1535 (2008).
Zimmerli, W. & Sendi, P. Orthopaedic biofilm infections. APMIS 125, 353–364 (2017).
Latha, T. et al. MRSA: the leading pathogen of orthopedic infection in a tertiary care hospital, South India. Afr. Health Sci. 19, 1393 (2019).
Klemt, C. et al. Single-Stage Revision of the Infected Total Knee Arthroplasty Is Associated With Improved Functional Outcomes: A Propensity Score–Matched Cohort Study. J. Arthroplast. 36, 298–304 (2021).
Massin, P. et al. Infection recurrence factors in one- and two-stage total knee prosthesis exchanges. Knee Surg. Sports Traumatol. Arthrosc. 24, 3131–3139 (2016).
Romanò, C. L., Gala, L., Logoluso, N., Romanò, D. & Drago, L. Two-stage revision of septic knee prosthesis with articulating knee spacers yields better infection eradication rate than one-stage or two-stage revision with static spacers. Knee Surg. Sports Traumatol., Arthrosc. 20, 2445–2453 (2012).
Löwik, C. A. M. et al. Debridement, antibiotics, and implant retention is a viable treatment option for early periprosthetic joint infection presenting more than 4 weeks after Index Arthroplasty. Clin. Infect. Dis. 71, 630–636 (2020).
Pangaud, C., Ollivier, M. & Argenson, J.-N. Outcome of single-stage versus two-stage exchange for revision knee arthroplasty for chronic periprosthetic infection. EFORT Open Rev. 4, 495–502 (2019).
Zhao, Y., Fan, S., Wang, Z., Yan, X. & Luo, H. Systematic review and meta-analysis of single-stage vs two-stage revision for periprosthetic joint infection: a call for a prospective randomized trial. BMC Musculoskelet. Disord. 25, 153 (2024).
Kunutsor, S. K., Whitehouse, M. R., Lenguerrand, E., Blom, A. W. & Beswick, A. D. Re-infection outcomes following one- and two-stage surgical revision of infected knee prosthesis: a systematic review and meta-analysis. PLoS One 11, e0151537 (2016).
Kaufman, M. G., Meaike, J. D. & Izaddoost, S. A. Orthopedic prosthetic infections: diagnosis and orthopedic salvage. Semin Plast. Surg. 30, 66–72 (2016).
Masters, E. A. et al. Skeletal infections: microbial pathogenesis, immunity and clinical management. Nat. Rev. Microbiol. 20, 385–400 (2022).
Garzoni, C. & Kelley, W. L. Return of the Trojan horse: intracellular phenotype switching and immune evasion by Staphylococcus aureus. EMBO Mol. Med. 3, 115–117 (2011).
Yu, B., Pacureanu, A., Olivier, C., Cloetens, P. & Peyrin, F. Assessment of the human bone lacuno-canalicular network at the nanoscale and impact of spatial resolution. Sci. Rep. 10, 4567 (2020).
Rong, Z. et al. Immune escape of Staphylococcus aureus mediated by osteocyte lacuna-canalicular network leads to persistent and uncured bone infection. Front. Cell Infect Microbiol. 15, 1592086 (2025).
Gleitz, H. F. E., Benabid, A. & Schneider, R. K. Still a burning question: the interplay between inflammation and fibrosis in myeloproliferative neoplasms. Curr. Opin. Hematol. 28, 364–371 (2021).
Katz, S. E. & Creech, C. B. Principles and Practice of Pediatric Infectious Diseases. https://doi.org/10.1016/C2019-0-00075-1. (Elsevier, 2023).
Narayanappa, G. & Nandeesh, B. N. Infective myositis. Brain Pathol. 31, e12950 (2021).
Oliveira, V. et al. Biofilm formation and expression of virulence genes of microorganisms grown in contact with a new bioactive glass. Pathogens 9, 927 (2020).
Rama Devi, K. et al. In vitro and in vivo efficacy of rosmarinic acid on quorum sensing mediated biofilm formation and virulence factor production in Aeromonas hydrophila. Biofouling 32, 1171–1183 (2016).
Antar, S. A., Ashour, N. A., Marawan, M. E. & Al-Karmalawy, A. A. Fibrosis: types, effects, markers, mechanisms for disease progression, and its relation with oxidative stress, immunity, and inflammation. Int J. Mol. Sci. 24, 4004 (2023).
Lieber, R. L. & Ward, S. R. Cellular Mechanisms of Tissue Fibrosis. 4. Structural and functional consequences of skeletal muscle fibrosis. Am. J. Physiol. -Cell Physiol. 305, C241–C252 (2013).
Smith, C. J. et al. Cold plasma generates a localized inflammatory response and promotes muscle repair. Adv. Ther. https://doi.org/10.1002/adtp.202500097 (2025).
Lee, T. C. et al. Detecting microdamage in bone. J. Anat. 203, 161–172 (2003).
Reikeras, O., Borgen, P., Reseland, J. E. & Lyngstadaas, S. P. Changes in serum cytokines in response to musculoskeletal surgical trauma. BMC Res. Notes 7, 128 (2014).
Takano, K. & Nakagawa, H. Contribution of cytokine-induced neutrophil chemoattractant CINC-2 and CINC-3 to neutrophil recruitment in lipopolysaccharide-induced inflammation in rats. Inflamm. Res. 50, 503–508 (2001).
Goh, Q. et al. Intercellular adhesion molecule-1 expression by skeletal muscle cells augments myogenesis. Exp. Cell Res 331, 292–308 (2015).
Lozano, D. D. L-Selectin and leukocyte function in skeletal muscle reperfusion injury. Arch. Surg. 134, 1079 (1999).
Cavalli, G. et al. Interleukin 1α: a comprehensive review on the role of IL-1α in the pathogenesis and treatment of autoimmune and inflammatory diseases. Autoimmun. Rev. 20, 102763 (2021).
Knight, B. E. et al. TIMP-1 attenuates the development of inflammatory pain through MMP-dependent and receptor-mediated cell signaling mechanisms. Front. Mol. Neurosci. 12, 220 (2019).
Dapunt, U., Maurer, S., Giese, T., Gaida, M. M. & Hänsch, G. M. The Macrophage Inflammatory Proteins MIP1 MIP1α (CCL3) and MIP2α (CXCL2) in implant-associated osteomyelitis: linking inflammation to bone degradation. Mediators Inflamm. 2014, 1–10 (2014).
Kassem, A., Lindholm, C. & Lerner, U. H. Toll-Like Receptor 2 Stimulation of Osteoblasts Mediates Staphylococcus Aureus Induced Bone Resorption and Osteoclastogenesis through Enhanced RANKL. PLoS One 11, e0156708 (2016).
Takegahara, N., Kim, H. & Choi, Y. Unraveling the intricacies of osteoclast differentiation and maturation: insight into novel therapeutic strategies for bone-destructive diseases. Exp. Mol. Med. 56, 264–272 (2024).
Dong, Q. et al. A review of bacterial and osteoclast differentiation in bone infection. Micro Pathog. 197, 107102 (2024).
Kim, M.-Y. et al. Function of CD4+CD3− cells in relation to B- and T-zone stroma in spleen. Blood 109, 1602–1610 (2007).
Lane, P. J. L., Gaspal, F. M. C. & Kim, M.-Y. Two sides of a cellular coin: CD4+CD3− cells regulate memory responses and lymph-node organization. Nat. Rev. Immunol. 5, 655–660 (2005).
Shi, Y. et al. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: what we do and don’t know. Cell Res 16, 126–133 (2006).
Liao, W., Lin, J.-X. & Leonard, W. J. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr. Opin. Immunol. 23, 598–604 (2011).
Ng, T. H. S. et al. Regulation of adaptive immunity: the role of Interleukin-10. Front. Immunol. 4, 129 (2013).
Wynn, T. A. IL-13 effector functions. Annu. Rev. Immunol. 21, 425–456 (2003).
Wu, Q., Tu, H. & Li, J. Multifaceted Roles of Chemokine C-X-C Motif Ligand 7 in Inflammatory Diseases and Cancer. Front. Pharmacol. 13, 914730 (2022).
Mahmoud, N. N. et al. Investigating Inflammatory Markers in Wound Healing: Understanding Implications and Identifying Artifacts. ACS Pharm. Transl. Sci. 7, 18–27 (2024).
Franzén, O., Gan, L.-M. & Björkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, baz046 (2019).
Castanza, A. S. et al. Extending support for mouse data in the Molecular Signatures Database (MSigDB). Nat. Methods 20, 1619–1620 (2023).
Li, K. et al. PD-1/PD-L1 blockade is a potent adjuvant in treatment of Staphylococcus aureus osteomyelitis in mice. Mol. Ther. 31, 174–192 (2023).
Urish, K. L. & Cassat, J. E. Staphylococcus aureus Osteomyelitis: Bone, Bugs, and Surgery. Infect. Immun. 88, e00932–19 (2020).
Croes, M. et al. Inflammation-Induced Osteogenesis in a Rabbit Tibia Model. Tissue Eng. C. Methods 23, 673–685 (2017).
Budin, M. et al. Comparison of patient-reported outcomes measures and quality-adjusted life years following one- and two-stage septic knee exchange. Antibiotics 11, 1602 (2022).
Wignadasan, W., Ibrahim, M. & Haddad, F. S. One- or two-stage reimplantation for infected total knee prosthesis? Orthop. Traumatology: Surg. Res. 109, 103453 (2023).
Pelt, C. E. et al. Two-stage revision TKA is associated with high complication and failure rates. Adv. Orthop. 2014, 1–7 (2014).
McLachlan, J. B. & Jenkins, M. K. Migration and accumulation of effector CD4+ T cells in nonlymphoid tissues. Proc. Am. Thorac. Soc. 4, 439–442 (2007).
Guo, M. et al. Molecular, metabolic, and functional CD4 T cell paralysis in the lymph node impedes tumor control. Cell Rep. 42, 113047 (2023).
Xue, J. et al. Lymph-derived chemokines direct early neutrophil infiltration in the lymph nodes upon Staphylococcus aureus skin infection. Proc. Natl. Acad. Sci. 119, e2111726119 (2022).
Wang, D., Wang, L., Liu, Q. & Zhao, Y. Virulence factors in biofilm formation and therapeutic strategies for Staphylococcus aureus: A review. Animals Zoonoses https://doi.org/10.1016/j.azn.2024.11.003 (2024).
Dastgheyb, S., Parvizi, J., Shapiro, I. M., Hickok, N. J. & Otto, M. Effect of Biofilms on Recalcitrance of Staphylococcal Joint Infection to Antibiotic Treatment. J. Infect. Dis. 211, 641–650 (2015).
Dastgheyb, S. S. et al. Role of Phenol-Soluble Modulins in Formation of Staphylococcus aureus Biofilms in Synovial Fluid. Infect. Immun. 83, 2966–2975 (2015).
Knott, S. et al. Staphylococcus aureus floating biofilm formation and phenotype in synovial fluid depends on albumin, fibrinogen, and hyaluronic acid. Front. Microbiol. 12, 655873 (2021).
de Jong, N. W. M., van Kessel, K. P. M. & van Strijp, J. A. G. Immune Evasion by Staphylococcus aureus. Microbiol. Spectr. 7, (2019).
Zhao, N. et al. Microbubble cavitation restores Staphylococcus aureus antibiotic susceptibility in vitro and in a septic arthritis model. Commun. Biol. 6, 425 (2023).
Shroff, K. E., Meslin, K. & Cebra, J. J. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 63, 3904–3913 (1995).
Madaro, L. & Bouché, M. From innate to adaptive immune response in muscular dystrophies and skeletal muscle regeneration: the role of lymphocytes. Biomed. Res Int 2014, 1–12 (2014).
Brady, R. A., Mocca, C. P., Plaut, R. D., Takeda, K. & Burns, D. L. Comparison of the immune response during acute and chronic Staphylococcus aureus infection. PLoS One 13, e0195342 (2018).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Todorov, V. & Filzmoser, P. An object-oriented framework for robust multivariate analysis. J. Stat. Softw. 32, 1–47 (2009).
Carlson, M. Genome wide annotation for Rat. Bioconductor 3.21.
Wickham, H. ggplot2: Elegant Graphics for Data Analysis. https://ggplot2.tidyverse.org (Springer-Verlag, New York).
Pedersen, T. patchwork: The Composer of Plots. R package version 1.3.0.9000, https://github.com/thomasp85/patchwork (2025).
Wickham, H., Pedersen, T. & Sidel, D. scales: Scale Functions for Visualization. R package version 1.4.0, https://scales.r-lib.org (2025).
Durinck, S., Spellman, P., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at https://doi.org/10.1101/060012 (2021).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 14, 7 (2013).
Hvitfeldt, E. Paletteer: Comprehensive Collection of Color Palettes. R package version 1.3.0 (2021).
Kolde, R. pheatmap: Pretty Heatmaps. R package version 1.0.13 (2025).
Wickham, H. forcats: Tools for Working with Categorical Variables (Factors). R package version 1.0.0, https://github.com/tidyverse/forcats (2023).
Wickham, H. stringr: Simple, Consistent Wrappers for Common String Operations. R package version 1.5.1, https://github.com/tidyverse/stringr (2023).
Acknowledgements
This study received support from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR076941). Histology funding was provided by a PCMD Admin Affiliate Member Award grant (P30AR069619) from the University of Pennsylvania Center for Musculoskeletal Disorders Histology Core. The funders played no role in study design, data collection, analysis, and interpretation of data, or the writing of this manuscript. Special thanks to Julie Engiles for help with histological staining and interpretation of histological results. Special thanks to Genevieve Baldomero for assistance with sample acquisition and processing. Special thanks to Edgardo Arroyo for help with IF staining and imaging. Special thanks to Christopher Talone, Jordan Safran, and Daya Rao for their help with micro-CT scanning and analysis. Special thanks to Brianna Nissley for help with histological scoring.
Author information
Authors and Affiliations
Contributions
C.J.S. data generation, data analysis, data interpretation and visualization, original manuscript drafting and revision. A.J.M., A.R.W., A.G.M., T.P.T., E.C., A.A.L. data generation, data analysis, data interpretation, manuscript revision, and editing. B.F.G., T.P.S., N.J.H. technical and methodological support, data interpretation, manuscript revisio,n and editing. T.A.F. data interpretation, technical and methodological support, inception of study idea, manuscript revision and editing, supervision, and funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Smith, C.J., Moniodes, A.J., Watkins, A.R. et al. Surgical revision in the presence of an S. aureus infection increases virulence factor expression and activates a multi-tissue inflammatory response. npj Biofilms Microbiomes 12, 18 (2026). https://doi.org/10.1038/s41522-025-00885-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-025-00885-6