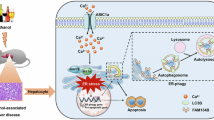
Abstract
Alcohol-associated liver disease (ALD), characterized by gut barrier disruption and microbial dysbiosis, is associated with significant depletion of the genus Bifidobacterium in patients, as evidenced by our cohort of 127 subjects. Functional screening revealed B. pseudocatenulatum as a protective strain. In a murine ALD model established with a Lieber–DeCarli ethanol diet, oral administration of B. pseudocatenulatum for 8 weeks ameliorated hepatomegaly, steatosis, and serum transaminase levels. Probiotic intervention restored intestinal barrier function, as indicated by reduced lipopolysaccharide-binding proteins and upregulated tight junction protein expression. Microbiome analysis revealed a mitigation of dysbiosis, with a reduction in pathogenic Escherichia-Shigella and Parabacteroides and an enrichment of beneficial Bifidobacterium and Blautia, concomitant with shifts in lipid metabolism. Mechanistically, B. pseudocatenulatum-derived short-chain fatty acids downregulated the expression of hepatic lipogenic genes (Cd36, Fasn, Accα) and pro-inflammatory cytokines (Il-1β, Ccl2, Tnf-α). These results suggest that B. pseudocatenulatum is a promising probiotic candidate for ALD management.
Similar content being viewed by others
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA009220), and are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
References
Seitz, H. K. et al. Publisher Correction: Alcoholic liver disease. Nat. Rev. Dis. Prim. 4, 18 (2018).
Patel, R. & Mueller, M. Alcoholic Liver Disease (StatPearls, 2022).
Wang, H., Mehal, W., Nagy, L. E. & Rotman, Y. Immunological mechanisms and therapeutic targets of fatty liver diseases. Cell Mol. Immunol. 18, 73–91 (2021).
Camilleri, M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut 68, 1516–1526 (2019).
Kapitan, M., Niemiec, M. J., Steimle, A., Frick, J. S. & Jacobsen, I. D. Fungi as part of the microbiota and interactions with intestinal bacteria. Curr. Top. Microbiol. Immunol. 422, 265–301 (2019).
Pohl, K., Moodley, P. & Dhanda, A. D. Alcohol’s impact on the gut and liver. Nutrients 13, 3170 (2021).
Wu, J. et al. Altered faecal microbiota on the expression of Th cells responses in the exacerbation of patients with hepatitis E infection. J. Viral Hepat. 27, 1243–1252 (2020).
Litwinowicz, K., Choroszy, M. & Waszczuk, E. Changes in the composition of the human intestinal microbiome in alcohol use disorder: a systematic review. Am. J. Drug Alcohol Abus. 46, 4–12 (2020).
Engen, P. A., Green, S. J., Voigt, R. M., Forsyth, C. B. & Keshavarzian, A. The gastrointestinal microbiome: alcohol effects on the composition of intestinal microbiota. Alcohol Res. 37, 223–236 (2015).
Bajaj, J. S. et al. Continued alcohol misuse in human cirrhosis is associated with an impaired gut-liver axis. Alcohol Clin. Exp. Res. 41, 1857–1865 (2017).
Llopis, M. et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 65, 830–839 (2016).
Ciocan, D. et al. Characterization of intestinal microbiota in alcoholic patients with and without alcoholic hepatitis or chronic alcoholic pancreatitis. Sci. Rep. 8, 4822 (2018).
Bajaj, J. S. et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 60, 940–947 (2014).
Wolfe, W. et al. The challenge of applications of probiotics in gastrointestinal diseases. Adv. Gut Microbiome Res. 2023, 1–10 (2023).
Nie, N. et al. Bifidobacterium plays a protective role in TNF-α-induced inflammatory response in Caco-2 cell through NF-κB and p38MAPK pathways. Mol. Cell. Biochem. 464, 83–91 (2020).
Cano, P. G., Santacruz, A., Trejo, F. M. & Sanz, Y. Bifidobacterium CECT 7765 improves metabolic and immunological alterations associated with obesity in high-fat diet-fed mice. Obesity 21, 2310–2321 (2013).
Chen, Y. et al. Bifidobacterium pseudocatenulatum ameliorates DSS-induced colitis by maintaining intestinal mechanical barrier, blocking proinflammatory cytokines, inhibiting TLR4/NF-κB signaling, and altering gut microbiota. J. Agric. Food Chem. 69, 1496–1512 (2021).
Mizokami, A., Kawakubo-Yasukochi, T. & Hirata, M. Osteocalcin and its endocrine functions. Biochem. Pharm. 132, 1–8 (2017).
Silva de Oliveira, R. et al. Bifidobacteria-derived exopolysaccharide promotes anti-tumor immunity. Cell Rep. 44, 116223 (2025).
Liu, L. et al. TWEAK-Fn14 signaling protects mice from pulmonary fibrosis by inhibiting fibroblast activation and recruiting pro-regenerative macrophages. Cell Rep. 44, 115220 (2025).
Liu, J. et al. TWEAK/Fn14 signals mediate burn wound repair. J. Invest. Dermatol. 139, 224–234 (2019).
Kawaratani, H. et al. The effect of inflammatory cytokines in alcoholic liver disease. Mediators Inflamm. 2013, 495156 (2013).
Muta, T. & Takeshige, K. Essential roles of CD14 and lipopolysaccharide-binding protein for activation of toll-like receptor (TLR)2 as well as TLR4 Reconstitution of TLR2- and TLR4-activation by distinguishable ligands in LPS preparations. Eur. J. Biochem. 268, 4580–4589 (2001).
Llorente, C. et al. mAChR4 suppresses liver disease via GAP-induced antimicrobial immunity. Nature 646, 180–189 (2025).
Jin, L. et al. Degradation products of polydopamine restrained inflammatory response of LPS-stimulated macrophages through mediation TLR-4-MYD88 dependent signaling pathways by antioxidant. Inflammation 42, 658–671 (2019).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Sarin, S. K., Pande, A. & Schnabl, B. Microbiome as a therapeutic target in alcohol-related liver disease. J. Hepatol. 70, 260–272 (2019).
Canesso, M. C. C. et al. Comparing the effects of acute alcohol consumption in germ-free and conventional mice: the role of the gut microbiota. BMC Microbiol. 14, 240 (2014).
Gao, B. & Bataller, R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 141, 1572–1585 (2011).
Fang, D. et al. Bifidobacterium pseudocatenulatum LI09 and Bifidobacterium catenulatum LI10 attenuate D-galactosamine-induced liver injury by modifying the gut microbiota. Sci. Rep. 7, 8770 (2017).
Keshavarzian, A. et al. Leaky gut in alcoholic cirrhosis: a possible mechanism for alcohol-induced liver damage. Am. J. Gastroenterol. 94, 200–207 (1999).
Choudhry, M. A., Fazal, N., Goto, M., Gamelli, R. L. & Sayeed, M. M. Gut-associated lymphoid T cell suppression enhances bacterial translocation in alcohol and burn injury. Am. J. Physiol. Gastrointest. Liver Physiol. 282, G937–G947 (2002).
Seki, E. et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat. Med. 13, 1324–1332 (2007).
Jiang, Y., Beller, D. I., Frendl, G. & Graves, D. T. Monocyte chemoattractant protein-1 regulates adhesion molecule expression and cytokine production in human monocytes. J. Immunol. 148, 2423–2428 (1992).
Sheron, N., Bird, G., Goka, J., Alexander, G. & Williams, R. Elevated plasma interleukin-6 and increased severity and mortality in alcoholic hepatitis. Clin. Exp. Immunol. 84, 449–453 (1991).
Gustot, T. et al. Differential liver sensitization to toll-like receptor pathways in mice with alcoholic fatty liver. Hepatology 43, 989–1000 (2006).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010).
Chassaing, B., Ley, R. E. & Gewirtz, A. T. Intestinal epithelial cell toll-like receptor 5 regulates the intestinal microbiota to prevent low-grade inflammation and metabolic syndrome in mice. Gastroenterology 147, 1363–1377.e1317 (2014).
Koblansky, A. A. et al. Recognition of profilin by Toll-like receptor 12 is critical for host resistance to Toxoplasma gondii. Immunity 38, 119–130 (2013).
Ji, C., Chan, C. & Kaplowitz, N. Predominant role of sterol response element binding proteins (SREBP) lipogenic pathways in hepatic steatosis in the murine intragastric ethanol feeding model. J. Hepatol. 45, 717–724 (2006).
Zhong, W. et al. Chronic alcohol exposure stimulates adipose tissue lipolysis in mice: role of reverse triglyceride transport in the pathogenesis of alcoholic steatosis. Am. J. Pathol. 180, 998–1007 (2012).
Li, L. C. et al. Palmitate aggravates proteinuria-induced cell death and inflammation via CD36-inflammasome axis in the proximal tubular cells of obese mice. Am. J. Physiol. Ren. Physiol. 315, F1720–F1731 (2018).
Arboleya, S., Watkins, C., Stanton, C. & Ross, R. P. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 7, 1204 (2016).
Shi, Z. et al. Anti-obesity effects of α-amylase inhibitor enriched-extract from white common beans (Phaseolus vulgaris L.) associated with the modulation of gut microbiota composition in high-fat diet-induced obese rats. Food Funct. 11, 1624–1634 (2020).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Backhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Rodriguez, J. et al. Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Gut 69, 1975–1987 (2020).
Agudelo-Ochoa, G. M. et al. Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes 12, 1707610 (2020).
Mancuso, C. & Santangelo, R. Alzheimer’s disease and gut microbiota modifications: the long way between preclinical studies and clinical evidence. Pharmacol. Res. 129, 329–336 (2018).
Huc, T., Nowinski, A., Drapala, A., Konopelski, P. & Ufnal, M. Indole and indoxyl sulfate, gut bacteria metabolites of tryptophan, change arterial blood pressure via peripheral and central mechanisms in rats. Pharmacol. Res. 130, 172–179 (2018).
Albillos, A., De Gottardi, A. & Rescigno, M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J. Hepatol. 72, 558–577 (2020).
Zaborska, K. E., Lee, S. A., Garribay, D., Cha, E. & Cummings, B. P. Deoxycholic acid supplementation impairs glucose homeostasis in mice. PLoS ONE 13, e0200908 (2018).
Astbury, S. et al. Lower gut microbiome diversity and higher abundance of proinflammatory genus Collinsella are associated with biopsy-proven nonalcoholic steatohepatitis. Gut Microbes 11, 569–580 (2020).
Grimble, R. F. et al. Cysteine and glycine supplementation modulate the metabolic response to tumor necrosis factor alpha in rats fed a low protein diet. J. Nutr. 122, 2066–2073 (1992).
Bajaj, J. S. et al. Fungal dysbiosis in cirrhosis. Gut 67, 1146–1154 (2018).
Forsyth, C. B. et al. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol 43, 163–172 (2009).
Cassard, A. M. & Ciocan, D. Microbiota, a key player in alcoholic liver disease. Clin. Mol. Hepatol. 24, 100–107 (2018).
Song, Q. et al. Bifidobacterium pseudolongum-generated acetate suppresses non-alcoholic fatty liver disease-associated hepatocellular carcinoma. J. Hepatol. 79, 1352–1365 (2023).
Ganesan, R. et al. Characteristics of microbiome-derived metabolomics according to the progression of alcoholic liver disease. Hepatol. Int. 18, 486–499 (2024).
Shu, X. et al. Bifidobacterium lactis TY-S01 protects against alcoholic liver injury in mice by regulating intestinal barrier function and gut microbiota. Heliyon 9, e17878 (2023).
Iwanaga, S. et al. Chromogenic substrates for horseshoe crab clotting enzyme. Its application for the assay of bacterial endotoxins. Haemostasis 7, 183–188 (1978).
Askgaard, G., Kraglund, F., Kann, A. E., Vilstrup, H. & Jepsen, P. [Epidemiology for alcohol-related liver disease]. Ugeskr. Laeger 183, V11200893 (2021).
Li, Y. et al. Bifidobacterium adolescentis CGMCC 15058 alleviates liver injury, enhances the intestinal barrier and modifies the gut microbiota in D-galactosamine-treated rats. Appl. Microbiol. Biotechnol. 103, 375–393 (2019).
Jiang, X. W. et al. New strain of Pediococcus pentosaceus alleviates ethanol-induced liver injury by modulating the gut microbiota and short-chain fatty acid metabolism. World J. Gastroenterol. 26, 6224–6240 (2020).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82502783); the Zhejiang Provincial Natural Science Foundation of China (LQ22H030013;2022-KY1-001-039); the Fundamental Research Funds for the Central Universities (2025ZFJH03); the State Key Laboratory for Diagnosis and Treatment of Infectious Disease Independent Project (zz202320); the Young Innovative Talents Support Program of Zhejiang Medical and Health Science and Technology Project (2022499572, 2022RC243); and the Young Innovative Talents Support Program of Zhejiang Chinese Medicine Science Technology Project (2024025437, 2024ZR152).
Author information
Authors and Affiliations
Contributions
Y.L. conceptualized the study design and experimental framework. L.Y., H.X. and X.B. conducted the experimental investigations and coordinated the specimen acquisition. W.W. performed nucleic acid library preparation and high-throughput sequencing and established analytical pipelines. D.S. spearheaded the computational analyses and bioinformatics interpretation. L.L. synthesized the experimental findings and composed the initial draft of the manuscript. All the authors provided final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Y., Yang, L., Xu, H. et al. Restoration of ethanol-induced Bifidobacterium pseudocatenulatum depletion ameliorates alcohol-associated liver disease. npj Biofilms Microbiomes (2026). https://doi.org/10.1038/s41522-026-00913-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-026-00913-z