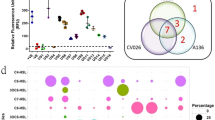
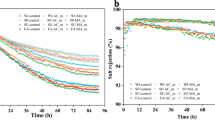
Abstract
Quorum sensing (QS) orchestrates collective microbial behaviors and functional acclimatization through chemical communication. However, QS in natural waters is challenged by dilution, alkaline hydrolysis, and enzymatic degradation of freely dissolved autoinducers. Here, we demonstrate that extracellular vesicles (EVs) act as selective, durable, and protective vectors for QS signal molecules under environmental stresses. Specifically, EVs preferentially package hydrophobic acyl‑homoserine lactones, concentrate them locally, and shield them from alkaline hydrolysis, and exhibiting long-distance transport. In addition, EVs possess specific affinity to recipients, thus influencing microbial community. Field investigation via multi-omics showed that EV abundance covaried with salinity, nutrients, chlorophyll a, and biomass, which were validated by culture experiments. Our statistical framework demonstrated that organisms producing moderate EV levels contributed significantly to maintaining community stability and ecosystem functions. Distinctively within this group, QS-active species (including Burkholderiaceae, Pseudomonadaceae, Rhodobacteraceae, Roseobacteraceae, Flavobacteriaceae etc.) emerge as key drivers facilitating these crucial ecological roles. Furthermore, metaproteomics of field EVs reveal QS receptor and synthesis proteins, suggesting coordinated transport of signals and proteins, which indicate new routes for QS crosstalk, particularly for taxa bearing luxR/I solos. Our results show that moderately generated EVs are the potentially important QS signal carriers and ecological regulation hubs in natural waters.
Similar content being viewed by others
Data availability
All the DNA sequences generated during the current study are available in the BIG Submission75, BIG Sub: https://ngdc.cncb.ac.cn/gsa/browse/CRA027247. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository76,77 with the dataset identifier PXD065377.
References
Mukherjee, S. & Bassler, B. L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382 (2019).
Coolahan, M. & Whalen, K. E. A review of quorum-sensing and its role in mediating interkingdom interactions in the ocean. Commun. Biol. 8, 179 (2025).
Decho, A. W., Norman, R. S. & Visscher, P. T. Quorum sensing in natural environments: emerging views from microbial mats. Trends Microbiol. 18, 73–80 (2010).
Papenfort, K. & Bassler, B. L. Quorum-sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588 (2016).
Abisado R.G. et al. Bacterial quorum sensing and microbial community interactions. mBio. 9, (2018).
Hmelo, L. R. Quorum sensing in marine microbial environments. Ann. Rev. Mar. Sci. 9, 257–281 (2017).
Horswill, A. R., Stoodley, P., Stewart, P. S. & Parsek, M. R. The effect of the chemical, biological, and physical environment on quorum sensing in structured microbial communities. Anal. Bioanal. Chem. 387, 371–380 (2007).
Wei, J. R. et al. A mobile quorum-sensing system in Serratia marcescens. J. Bacteriol. 188, 1518–1525 (2006).
Yates, E. A. et al. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70, 5635–5646 (2002).
Wellington, S. & Greenberg, E. P. Quorum sensing signal selectivity and the potential for interspecies cross talk. mBio 10, e00146–00119 (2019).
Slinger, B. L., Banerjee, S., Chandler, J. R. & Blackwell, H. E. Interspecies crosstalk via LuxI/LuxR-type quorum sensing pathways contributes to decreased nematode survival in coinfections of Pseudomonas aeruginosa and Burkholderia multivorans. ACS Chem. Biol. 19, 2557–2568 (2024).
Wu, F., Menn, D. J. & Wang, X. Quorum-sensing crosstalk-driven synthetic circuits: from unimodality to trimodality. Chem. Biol. 21, 1629–1638 (2014).
Arashida, N. et al. Identification of novel long chain N-acylhomoserine lactones of chain length C(20) from the marine phototrophic bacterium Rhodovulum sulfidophilum. Biosci. Biotechnol. Biochem. 82, 1683–1693 (2018).
Decho, A. W. et al. Autoinducers extracted from microbial mats reveal a surprising diversity of N-acylhomoserine lactones (AHLs) and abundance changes that may relate to diel pH. Environ. Microbiol. 11, 409–420 (2009).
Wang, Y. C. et al. Diversity, influential factor, and communication network construction of quorum-sensing bacteria in global wastewater treatment plants. Water Res. 279, 123437 (2025).
Wu, S. et al. Machine learning aided construction of the quorum-sensing communication network for human gut microbiota. Nat. Commun. 13, 3079 (2022).
Albataineh, H., Duke, M., Misra, S. K., Sharp, J. S. & Stevens, D. C. Identification of a solo acylhomoserine lactone synthase from the myxobacterium Archangium gephyra. Sci. Rep. 11, 3018 (2021).
Fuqua, C. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J. Bacteriol. 188, 3169–3171 (2006).
Roche, D. M. et al. Communications blackout? Do N-acylhomoserine-lactone-degrading enzymes have any role in quorum sensing? Microbiology 150, 2023–2028 (2004).
Hudaiberdiev, S. et al. Census of solo LuxR genes in prokaryotic genomes. Front. Cell Infect. Microbiol. 5, 20 (2015).
Zan, J. et al. A solo luxI-type gene directs acylhomoserine lactone synthesis and contributes to motility control in the marine sponge symbiont Ruegeria sp. KLH11. Microbiology 161, 50–56 (2015).
Chandler, J. R., Heilmann, S., Mittler, J. E. & Greenberg, E. P. Acyl-homoserine lactone-dependent eavesdropping promotes competition in a laboratory co-culture model. ISME J. 6, 2219–2228 (2012).
Toyofuku, M., Schild, S., Kaparakis-Liaskos, M. & Eberl, L. Composition and functions of bacterial membrane vesicles. Nat. Rev. Microbiol. 21, 415–430 (2023).
Zhu, L. T. et al. Diverse functional genes harboured in extracellular vesicles from environmental and human microbiota. J. Extracell. Vesicles 11, e12292 (2022).
Biller, S. J. et al. Bacterial vesicles in marine ecosystems. Science 343, 183–186 (2014).
Qin, Y. et al. Widespread of potential pathogen-derived extracellular vesicles carrying antibiotic resistance genes in indoor dust. Environ. Sci. Technol. 56, 5653–5663 (2022).
Long, L. et al. Extracellular vesicles are prevalent and effective carriers of environmental allergens in indoor dust. Environ. Sci. Technol. 59, 1969–1983 (2025).
Teng, Y. et al. Plant-derived exosomal micrornas shape the gut microbiota. Cell Host Microbe 24, 637–652.e638 (2018).
Gaudin, M. et al. Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ. Microbiol. Rep. 5, 109–116 (2013).
Wang, Z. et al. Fungal extracellular vesicles mediate cross-kingdom trafficking of virulence effectors into plant cells to promote infection. Mol. Plant 18, 1369–1389 (2025).
Biller, S. J. et al. Prochlorococcus extracellular vesicles: molecular composition and adsorption to diverse microbes. Environ. Microbiol. 24, 420–435 (2022).
Hackl, T. et al. Novel integrative elements and genomic plasticity in ocean ecosystems. Cell 186, 47–62.e16 (2023).
Toyofuku, M. et al. Membrane vesicle-mediated bacterial communication. ISME J. 11, 1504–1509 (2017).
Mashburn, L. M. & Whiteley, M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437, 422–425 (2005).
Emge, P. et al. Resilience of bacterial quorum sensing against fluid flow. Sci. Rep. 6, 33115 (2016).
Wang, J. et al. AHLs-mediated quorum sensing threshold and its response towards initial adhesion of wastewater biofilms. Water Res. 194, 116925 (2021).
Zhu, Y. L., Hou, H. M., Zhang, G. L., Wang, Y. F. & Hao, H. S. AHLs regulate biofilm formation and swimming motility of Hafnia alvei H4. Front Microbiol. 10, 1330 (2019).
Chu, W., Vattem, D. A., Maitin, V., Barnes, M. B. & McLean, R. J. Bioassays of quorum sensing compounds using Agrobacterium tumefaciens and Chromobacterium violaceum. Methods Mol. Biol. 692, 3–19 (2011).
Agmo Hernández V. An overview of surface forces and the DLVO theory. ChemTexts. 9 (2023).
Hmelo, L. & Van Mooy, B. A. S. Kinetic constraints on acylated homoserine lactone-based quorum sensing in marine environments. Aquat. Microb. Ecol. 54, 127–133 (2009).
Pearson, J. P., Delden, C. V. & Iglewski, B. H. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181, 1203–1210 (1999).
Chan, Y. Y. et al. Control of quorum sensing by a Burkholderia pseudomallei multidrug efflux pump. J. Bacteriol. 189, 4320–4324 (2007).
Buroni, S. et al. Assessment of three Resistance-Nodulation-Cell Division drug efflux transporters of Burkholderia cenocepacia in intrinsic antibiotic resistance. BMC Microbiol. 9, 200 (2009).
Gantner, S. et al. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 56, 188–194 (2006).
MacDonald, K. L. & Beveridge, T. J. Bactericidal effect of gentamicin-induced membrane vesicles derived from Pseudomonas aeruginosa PAO1 on gram-positive bacteria. Can. J. Microbiol. 48, 810–820 (2002).
Tashiro, Y. et al. Interaction of bacterial membrane vesicles with specific species and their potential for delivery to target cells. Front. Microbiol. 8, 571 (2017).
Schatz, D. et al. Communication via extracellular vesicles enhances viral infection of a cosmopolitan alga. Nat. Microbiol. 2, 1485–1492 (2017).
Case, R. J., Labbate, M. & Kjelleberg, S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2, 345–349 (2008).
Monnet, V. & Gardan, R. Quorum-sensing regulators in Gram-positive bacteria: ‘cherchez le peptide. Mol. Microbiol. 97, 181–184 (2015).
Lequette, Y., Lee, J. H., Ledgham, F., Lazdunski, A. & Greenberg, E. P. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 188, 3365–3370 (2006).
McIntosh, M., Krol, E. & Becker, A. Competitive and cooperative effects in quorum-sensing-regulated galactoglucan biosynthesis in Sinorhizobium meliloti. J. Bacteriol. 190, 5308–5317 (2008).
Subramoni, S. & Venturi, V. LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 155, 1377–1385 (2009).
Morris JJ, Lenski RE, Zinser ER. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio. 3, (2012).
Lerat, E. & Moran, N. A. The evolutionary history of quorum-sensing systems in bacteria. Mol. Biol. Evol. 21, 903–913 (2004).
Rice EW, Baird RB, Eaton AD. Standard Methods for the Examination of Water and Wastewater, 23rd Edition (2017).
Lv, H. et al. Temperature and nutrients are significant drivers of seasonal shift in phytoplankton community from a drinking water reservoir, subtropical China. Environ. Sci. Pollut. Res. Int. 21, 5917–5928 (2014).
He, W., Yang, H., Wang, X., Li, H. & Dong, Q. Growth of Salmonella Enteritidis in the presence of quorum sensing signaling compounds produced by Pseudomonas aeruginosa. Int. J. Food Eng. 17, 959–968 (2021).
Fletcher, M. P., Diggle, S. P., Camara, M. & Williams, P. Biosensor-based assays for PQS, HHQ and related 2-alkyl-4-quinolone quorum-sensing signal molecules. Nat. Protoc. 2, 1254–1262 (2007).
Ziegler, E. W., Brown, A. B., Nesnas, N. & Palmer, A. G. Abiotic hydrolysis kinetics of N-Acyl-L-homoserine Lactones: natural silencing of bacterial quorum sensing signals. Eur. J. Org. Chem. 2019, 2850–2856 (2019).
Huang, Y. L., Ki, J. S., Lee, O. O. & Qian, P. Y. Evidence for the dynamics of Acyl homoserine lactone and AHL-producing bacteria during subtidal biofilm formation. ISME J. 3, 296–304 (2009).
Mo, Y. et al. Low shifts in salinity determined assembly processes and network stability of microeukaryotic plankton communities in a subtropical urban reservoir. Microbiome 9, 128 (2021).
Tkacz, A., Hortala, M. & Poole, P. S. Absolute quantitation of microbiota abundance in environmental samples. Microbiome 6, 110 (2018).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017).
Uritskiy, G. V., DiRuggiero, J. & Taylor, J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 158 (2018).
Yuan, M. M. et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11, 343–348 (2021).
Rosseel, Y. Lavaan: an R package for structural equation modeling. J. Stat. Software 48, 1–36 (2012).
Kenny, D. A., Kaniskan, B. & McCoach, D. B. The performance of RMSEA in models with small degrees of freedom. Sociol. Methods Res. 44, 486–507 (2014).
JL Z, KP M. spaa: an R package for computing species association and niche overlap. Research Progress of Biodiversity Conservation in China. 165–174 (2014).
Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, (2011).
Kleyer, M. et al. Assessing species and community functional responses to environmental gradients: which multivariate methods? J. Veg. Sci. 23, 805–821 (2012).
Lai, J., Zou, Y., Zhang, J. & Peres-Neto, P. R. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca.hp R package. Methods Ecol. Evol. 13, 782–788 (2022).
Peng C, Chen Q, Tan S, Shen X, Jiang C. Generalized reporter score-based enrichment analysis for omics data. Brief Bioinform. 25, bbae116 (2024).
Shenhav, L. et al. FEAST: fast expectation-maximization for microbial source tracking. Nat. Methods 16, 627–632 (2019).
Ofek-Lalzar, M. et al. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 5, 4950 (2014).
Members & Partners, C.-N. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2025. Nucleic Acids Res. 53, D30–D44 (2025).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217 (2019).
Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 50, D1522–D1527 (2022).
Acknowledgements
This work was supported by National Natural Science Foundation of China (32161143016, 42177362), and National Basic Science Data Center “Environment Health DataBase” (NO. NBSDC-DB-21). We thank Prof. Weihua Chu from China Pharmaceutical University for his help in QS biosensor experiments. We thank Prof. Steven Biller for his advice and assistance on this paper.
Author information
Authors and Affiliations
Contributions
X.L.X. conducted the experiments, generated and analyzed the data, and wrote the manuscript. J.J.L. performed the microcosmic experiments. L.T.Z. helped with the data analysis and revised the manuscript. L.L. and Y.F.D. provided the suggestions for computational adjustments. L.F.L. and J.H. contributed to the AHLs testing experiments. H.H.C. helped with measuring environmental factors. Q.S.H. designed the experiments, wrote the manuscript, and got the funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, X., Lin, J., Zhu, LT. et al. Extracellular vesicles as structured vectors of quorum sensing signals influence aquatic microbial communities. npj Biofilms Microbiomes (2026). https://doi.org/10.1038/s41522-026-00924-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-026-00924-w