Abstract
UBR5 encodes an E3 ubiquitin-protein ligase which targets distinct N-terminal residues of proteins for degradation. Heterozygous loss-of-function variants were reported in patients with Autism Spectrum Disorder (ASD) and developmental delay, and recently in a cohort of individuals with neurodevelopmental disorders and variable other features. Here, we report three unrelated individuals with de novo loss-of-function variants in UBR5, presenting with ASD and intellectual disability. We review the literature for other de novo predicted loss-of-function variants in probands with ASD or developmental delay (in total n = 11 variants), providing further evidence that UBR5 haploinsufficiency is associated with ASD and atypical neurodevelopmental trajectories, including developmental delay and intellectual disability.
Similar content being viewed by others
Introduction
Ubiquitination describes the attachment of ubiquitin to proteins, which regulates many aspects of protein function and stability, including protein degradation via the proteasome, subcellular localization, and interactions with other molecules. Ubiquitin protein ligase E3 component n-recognin 5 (UBR5), a gene mapping to chromosome 8q22, encodes an E3 ubiquitin-protein ligase with substrate specificity. It is expressed in the brain and targets distinct N-terminal residues of proteins. UBR5 has primarily been studied for its role in cancer, but also has essential functions for early development1. UBR5 is crucial for embryonic stem cell growth and maintenance of pluripotency2,3. Heterozygous Ubr5 mouse knockouts were reported to have normal development and fertility, whereas homozygous knockout mice were embryonically lethal, with delayed growth and development including impaired extraembryonic vascular development4. A definitive biological role or list of target genes and cellular pathways have yet to be established.
In humans, loss-of-function (LOF) variants in UBR5 are exceptionally rare in the general population (gnomAD pLI score 1), indicating negative selection5. Heterozygous de novo LOF variants in UBR5 were reported in cohorts of Autism Spectrum Disorder (ASD) and intellectual disability (ID)6,7,8,9,10,11. In addition, a recent study described 9 individuals with 8 different LOF variants and 8 individuals with 7 missense/in-frame variants that resulted in reduced ubiquitination activity or altered subcellular location. All variants were either de novo or found in mosaic state in a parent12. However, the gene-disease association remains to be further characterized. Other genes encoding ubiquitin-protein ligases within this N-end rule pathway (UBR1, UBR6, and UBR7) have been associated with autosomal recessive or autosomal dominant neurodevelopmental syndromes. UBR1 deficiency causes autosomal recessive Johanson-Blizzard syndrome (OMIM-P 243800), and biallelic variants in UBR7 are associated with autosomal recessive Li-Campeau syndrome (OMIM-P 619189). Haploinsufficiency of UBR6, currently known as FBXO11, is associated with autosomal dominant intellectual disability, distinctive facial features, and behavioral abnormalities (including ASD) (OMIM-P 618089).
Here, we describe three unrelated individuals with de novo, predicted LOF variants in UBR5 from the Autism Speaks MSSNG whole genome sequencing collection6. All individuals presented with ASD and ID. We also review previously reported de novo LOF variants from the literature and other data repositories and provide additional evidence that UBR5 haploinsufficiency is associated with variable neurodevelopmental phenotypes, including ASD, developmental delay and ID.
Results
MSSNG cohort
Three individuals with de novo, predicted loss-of-function variants in UBR5 presented with ASD and ID. No other pathogenic or likely pathogenic genomic variants in neurodevelopmental disease genes were identified by genome sequencing in these three individuals.
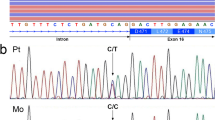
Individual 1 (AU2562301) has ASD and ID, and was identified with a de novo pathogenic deletion of 4 nucleotides in UBR5, predicted to result in a frameshift variant (NM_015902.6(UBR5): c.4447_4450del; p.Ile1483Phefs*21). The first developmental and behavioral concerns became evident at 2.5 years of age. He walked unaided at the age of 12 months and spoke first words at the age of 48 months. His growth parameters were within normal limits with a head circumference in the upper normal range (Z-score at 9 years: height -0.74; weight -0.32; and head circumference 1.81; at 17 years: height -1.45, and head circumference -0.45). Cognitive developmental testing administered between 7 and 10 years showed extremely low cognitive functioning (full scale IQ score, verbal IQ score, and performance IQ score all in the extremely low range on the Griffiths) and extremely low adaptive functioning (socialization, communication, daily living skills, and adaptive behavior domains all in the extremely low range on the Vineland Adaptive Behavior Scales 1984 Edition). No childhood regression or loss of skills were reported. At 17 years of age, he exhibited no signs of epilepsy, movement disorders, or other neurological abnormalities, and brain MRI findings were unremarkable. Behavioral concerns included anxiety and psychomotor agitation. Facial features were notable for prominent ears.
Individual 2 (AU2618301) was identified with a de novo pathogenic deletion of two nucleotides in UBR5, predicted to result in a frameshift variant (NM_015902.6(UBR5): c.752_753del; p.Leu251Profs*2). He was diagnosed with ASD and significant language development disorder. Growth measurements taken at 9 years showed height in the upper normal range and macrocephaly (Z-score at 9 years: height 1.71; head circumference 3.56). Cognitive developmental testing administered between 4 and 5 years showed extremely low cognitive functioning (non-verbal IQ score in the extremely low range on the Raven version 1) and extremely low adaptive functioning (socialization, communication, daily living skills, adaptive behaviour, and motor skills domains all in the extremely low range on the Vineland Adaptive Behavior Scales 1984 Edition). Autistic features included impairments in communication and social interaction, as well as repetitive, restrictive, and stereotyped behaviors, as observed on the ADOS at age 9. At age 4, the ADI-R similarly indicated delays in communication and language, difficulties in social interaction, repetitive and stereotyped behaviors, and abnormal early development. He did not present with epilepsy, movement disorders, other neurological abnormalities, congenital anomalies, or distinctive facial features. A brain MRI performed at age 4 revealed marked bifrontal underdevelopment of the cerebral hemispheres and streaky hyperintensities in the white matter.
Individual 3 (7-0657-003) was identified with a pathogenic de novo canonical splice site variant in UBR5, predicted to change the acceptor site 2 bps downstream (NM_015902.6(UBR5): c.4059-2 A > G; p.?). He began crawling at 9 months, walked unaided at 18 months, and spoke first words at 18 months, 2-3 word phrases at 2 years, and full sentences at 3-4 years. Developmental and behavioral concerns emerged around age 3, including globally slow learning, delayed language and pre-academic skills, limited peer interactions, absence of imaginative play, and anxiety. He was subsequently diagnosed with ASD (based on ADOS-2, CARS-2, structured parent interview, and direct observation by a licensed psychologist). Additional diagnoses included Attention-Deficit/Hyperactivity Disorder (ADHD) and sensory processing disorder. Delays were noted across expressive and receptive language, fine motor skills, sensory processing, and activities of daily living. Growth parameters at 3 years were within normal limits (Z-score at 3 years: height −0.59; weight −0.28). Physical examination was unremarkable, including normal findings across head, eyes, ENT, neck, cardiovascular, gastrointestinal, integumentary, genitourinary, lymphatic, musculoskeletal, skin, and neurologic systems. At age 10, cognitive testing revealed very low non-verbal IQ on the Stanford-Binet Intelligence Scales (Fifth Edition) and extremely low scores on the Adaptive Behavior Assessment System (Third Edition). By age 13, he was enrolled in a special education classroom, focusing on daily living, self-care, pre-vocational skills, and fostering independence. Caregivers reported behavioral challenges, including outbursts triggered by frustration and sound sensitivity.
Published cohorts
We searched the medical and scientific literature for additional loss-of-function variants in UBR5 and identified another 8 loss-of-function variants in 9 individuals with ASD and/or developmental disorders, in addition to the recent report of 15 LOF and functionally confirmed missense/in-frame variants by Sabeh et al. (Table 1). Five of the 9 individuals were from ASD cohorts, while four were recruited for developmental delay as their primary phenotype. Two individuals had IQ data available and had scores in the very low-borderline impaired range (Leiter-R7,) and low average range (testing method unavailable9,10. No UBR5 single gene deletions were reported so far, however, larger ( > 1 Mb) 8q22.2-q22.3 deletions encompassing UBR5 and multiple other OMIM genes are associated with intellectual disability, ASD, and seizures13,14,15.
Gene evidence
De novo UBR5 loss-of-function variants in individuals with ASD, ID and/or developmental delay (DD) were distributed throughout the gene with no obvious clustering (Fig. 1). Loss-of-function variants in UBR5 are overall very rare in the gnomAD v4.1.0 dataset, indicating a high probability of being loss-of-function intolerant (gnomAD pLI score 1, and a low observed/expected (oe) metric of 0.05 (0.03–0.07)). No inherited deletions of the gene were reported in the Database of Genomic Variants (DGV)16 or Decipher15.
Putative structure of UBR5 (NM_015902.6) consists of ubiquitin-associated (UBA) domain, Zinc finger domain, poly(A)-binding protein C-terminal domain (PABC), and HECT domain (https://www.uniprot.org/). Location of predicted loss-of-function variants (above) and missense/in-frame variants with functional evidence (below). Genomic size: 160,428 nucleotides, exon count: 59, Protein: 2799 amino acids, 18,734 - 158,954 nucleotides (multiple isoforms spanning 12–59 exons).
UBR5 also has high missense constraint scores (z = 8.38; o/e = 0.61; 0.59–0.63), indicating that missense variants are also under negative selective pressure. De novo missense variants of uncertain significance with allele frequencies = 0 in gnomAD were reported in additional individuals from MSSNG and other cohorts (Table S1, Fig. S1), however their functional and clinical significance remained unconfirmed.
Discussion
We describe three unrelated individuals with de novo predicted LOF variants in UBR5 (2 frameshift, 1 splice site). These findings are consistent with other variants described in the literature, including 23 de novo LOF and functionally confirmed missense/in-frame variants in individuals with ASD, intellectual disability, or developmental delay7,8,9,10,11,12,17. Variants were mostly unique per family and were distributed throughout the gene. By their location, all LOF variants were predicted to cause nonsense-mediated mRNA decay resulting in reduced gene expression18. Most of the individuals were described in the context of large cohort studies, some with limited phenotype information. However, the frequent reporting of de novo occurrence in this and the previous study, and high loss-of-function constraint metrics (pLI of 1 in the latest gnomAD release v.4.1.0), support a pathogenic impact of UBR5 haploinsufficiency.
Evaluation of Autism Gene Link Evidence (EAGLE) is a multi-disciplinary consensus-based scoring system for autism-associated genes19. A high curation score of 18.45 provided strong evidence for UBR5 being linked to ASD (https://gene.sfari.org/database/human-gene/UBR5). Most (7/11) of the de novo loss-of-function variants in UBR5 in this study were identified in ASD cohorts, suggesting that ASD is a highly prevalent phenotype. Other neurodevelopmental features, including intellectual disability and developmental delay, were frequently reported (Table 1)12.Atypical language development and low verbal IQ scores were reported, though some individuals demonstrated average performance or non-verbal IQ scores. In the study by Sabeh et al., heterozygous UBR5 loss-of-function variants were not associated with a consistent pattern of physical or growth abnormalities; however, a subset of individuals presented with features such as short or tall stature, microcephaly or macrocephaly, epilepsy, movement disorders, and cardiac or genital anomalies. Of the three additional probands with de novo loss-of-function variants in UBR5 reported in this study, one individual presented with abnormal brain MRI findings and macrocephaly. Although clinical variability appears to be substantial, heterogeneous phenotypic data may limit accurate comparisons, and some of the reported features could be attributable to alternative, including multifactorial, etiologies. Heterozygous mouse and zebrafish models did not reveal obvious morphological abnormalities4,20, and effects of UBR5 loss on gene expression were suggested to become more apparent at later developmental stages and potentially confined to specific tissues21. While the gene is widely expressed across various tissues, the clinical manifestations associated with haploinsufficiency remain to be fully characterized.
UBR5 encodes an E3 ubiquitin-protein ligase which targets distinct N-terminal residues of proteins for degradation. UBR-5 was suggested to upregulate SWI/SNF levels in C. elegans and may thus regulate various developmental processes21. Alterations affecting other ubiquitin-protein ligases with N-terminal substrate specificity (UBR1 and UBR7) have been associated with rare autosomal recessive neurodevelopmental syndromes: Biallelic loss of UBR1 causes Johanson-Blizzard syndrome, which is characterized by variable intellectual impairment, short stature, failure to thrive, exocrine pancreatic insufficiency, cardiac anomalies, hypothyroidism, typical facial features, and genital anomalies (OMIM-P 243800). Biallelic loss of UBR7 causes Li-Campeau syndrome. Patients with this condition can present with developmental delay, intellectual disability, epilepsy, hypotonia, short stature, cardiac anomalies, and hypothyroidism (OMIM-P 619189). UBR5 was suggested to have overlapping functions with UBR7 in C. elegans22, however, no biallelic loss-of-function variants in UBR5 have so far been reported in humans, and mouse knockouts are embryonically lethal4. Haploinsufficiency of UBR6, currently known as FBXO11, is associated with an autosomal dominant neurodevelopmental disorder, characterized by intellectual disability, distinctive facial features, and behavioral abnormalities (including ASD) (OMIM-P 618089). For UBR4, there is supporting evidence that suggests that the gene might be linked to episodic ataxia23,24,25. However, this association requires further confirmatory evidence.
Seven de novo missense/in-frame variants in UBR5 were reported to impair in vitro autoubiquitination or alter subcellular localization (Fig. 1 and Table 1)12. Additional de novo missense variants in UBR5 and associated phenotypes are listed (Table S1). Although de novo occurrence of a genomic variant may support its potential pathogenicity, it is not sufficient on its own to establish a pathogenic classification, particularly when the associated phenotype is nonspecific and genetically heterogeneous, as is often the case with neurodevelopmental disorders. Platforms such as GeneMatcher are valuable for facilitating collaboration among researchers and clinicians studying the same genes. However, the non-systematic nature of data submission and the overrepresentation of phenotypically similar cases can introduce confirmation bias, particularly in the context of de novo variants in genes considered “of interest”. In the study by Sabeh et al., most of the experimentally assessed variants did not clearly localize to known functional domains of the UBR5 protein, aside from two variants within the zinc finger domain (Fig. 1). This limits the ability to draw conclusions about the pathogenicity of other de novo missense variants in UBR5 in the absence of functional validation.
Heterozygous loss-of-function variants in UBR5 are associated with autism, developmental delay, and impaired cognitive functioning ranging from extremely low to borderline. While our data further support an important function of UBR5 on neurodevelopment, the molecular mechanisms and the spectrum of clinical presentations, including potential genotype-phenotype correlations, remain to be characterized further.
Methods
MSSNG cohort
For MSSNG probands, whole genome sequencing on whole blood or saliva was performed at The Centre for Applied Genomics (TCAG) in Toronto, Canada, as described previously6. Fragmented DNA (average 350 bp) was end-repaired, A-tailed and ligated with TruSeq Illumina adapters prior to library amplification. Validated libraries were pooled in equimolar quantities and sequenced on a HiSeq X platform or NovaSeq 6000 (Illumina, CA, USA) following the manufacturer’s protocol to generate paired-end reads of 150 bases in length. Reads were mapped using the Burrows-Wheeler Aligner (BWA). Single-nucleotide variants and small insertions/deletions were called using GATK. Copy number variants (CNVs) were called using a modified read depth method with the programs Estimation by Read Depth with Single-nucleotide variants and CNVnator using a window size of 500 bp17. Variant calls were annotated using a custom pipeline developed at TCAG based on ANNOVAR. PCR and Sanger sequencing were performed for validation and co-segregation analyses. Primer pairs are available upon request. All variant coordinates refer to the hg38/GRCh38 human reference genome.
Literature review
After identifying three de novo loss-of-function variants in individuals with ASD and ID, we performed a literature review and identified another 9 individuals with 8 different de novo loss-of-function variants in UBR5. Including the recently published cohort by Sabeh et al. 202512, there is a total of 26 unique variants in UBR5 reported to date. Those individuals are part of SFARI datasets (including the Simons Simplex Collection (SSC)26 and SPARK27), the Autism Sequencing Consortium (ASC), and other international cohorts of autism and developmental disorders.
Phenotypic data was extracted from study records (MSSNG) or original manuscripts (literature review).
Ethics statement
This study complied with all relevant ethical regulations including the Declaration of Helsinki. Informed consent was obtained from Autism Speaks MSSNG participants, and the study was approved by the Research Ethics Board at The Hospital for Sick Children.
Data availability
Access to the genome sequence and phenotype information from MSSNG can be obtained by completing data access agreements (https://research.mss.ng), as was done for this study.
References
Shearer, R. F., Iconomou, M., Watts, C. K. W. & Saunders, D. N. Functional roles of the E3 ubiquitin ligase UBR5 in cancer. Mol. Cancer Res. 13, 1523–1532 (2015).
Plank, M. et al. An analysis and validation pipeline for large-scale RNAi-based screens. Sci. Rep. 3, 1076 (2013).
Buckley, S. M. et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 11, 783–798 (2012).
Saunders, D. N. et al. Edd, the murine hyperplastic disc gene, is essential for yolk sac vascularization and chorioallantoic fusion. Mol. Cell Biol. 24, 7225–7234 (2004).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Trost, B. et al. Genomic architecture of autism from comprehensive whole-genome sequence annotation. Cell 185, 4409–4427.e18 (2022).
Viggiano, M. et al. Genomic analysis of 116 autism families strengthens known risk genes and highlights promising candidates. NPJ Genom. Med. 9, 21 (2024).
Fu, J. M. et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 54, 1320–1331 (2022).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
Kaplanis, J. et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature 586, 757–762 (2020).
Sabeh, P. et al. Heterozygous UBR5 variants result in a neurodevelopmental syndrome with developmental delay, autism, and intellectual disability. Am. J. Hum. Genet. 112, 75–86 (2025).
Kuechler, A. et al. Five patients with novel overlapping interstitial deletions in 8q22.2q22.3. Am. J. Med. Genet. A 155A, 1857–1864 (2011).
Kuroda, Y. et al. Refinement of the deletion in 8q22.2-q22.3: the minimum deletion size at 8q22.3 related to intellectual disability and epilepsy. Am. J. Med. Genet. A 164A, 2104–2108 (2014).
Firth, H. V. et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533 (2009).
MacDonald, J. R., Ziman, R., Yuen, R. K. C., Feuk, L. & Scherer, S. W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 42, D986–D992 (2014).
Trost, B. et al. A comprehensive workflow for read depth-based identification of copy-number variation from whole-genome sequence data. Am. J. Hum. Genet. 102, 142–155 (2018).
Schoenberg, D. R. & Maquat, L. E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 13, 246–259 (2012).
Schaaf, C. P. et al. A framework for an evidence-based gene list relevant to autism spectrum disorder. Nat. Rev. Genet. 21, 367–376 (2020).
LaCoursiere, C. M. et al. Zebrafish models of candidate human epilepsy-associated genes provide evidence of hyperexcitability. iScience. 27, 110172 (2024).
Lampersberger, L. et al. Loss of the E3 ubiquitin ligases UBR-5 or HECD-1 restores Caenorhabditis elegans development in the absence of SWI/SNF function. Proc. Natl. Acad. Sci. USA 120, e2217992120 (2023).
Li, C. et al. UBR7 functions with UBR5 in the Notch signaling pathway and is involved in a neurodevelopmental syndrome with epilepsy, ptosis, and hypothyroidism. Am. J. Hum. Genet. 108, 134–147 (2021).
Conroy, J. et al. A novel locus for episodic ataxia: UBR4 the likely candidate. Eur. J. Hum. Genet. 22, 505–510 (2014).
Choi, K.-D. et al. Genetic variants associated with episodic ataxia in Korea. Sci. Rep. 7, 13855 (2017).
Monies, D. et al. Lessons learned from large-scale, first-tier clinical exome sequencing in a highly consanguineous population. Am. J. Hum. Genet. 104, 1182–1201 (2019).
Fischbach, G. D. & Lord, C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 68, 192–195 (2010).
The SPARK Consortium. SPARK: a US cohort of 50,000 families to accelerate autism research. Neuron 97, 488–493 (2018).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17, 405–424 (2015).
Acknowledgements
The authors wish to acknowledge the resources of MSSNG (http://www.mss.ng/), Autism Speaks and The Centre for Applied Genomics at The Hospital for Sick Children, Toronto, Canada. We also thank the participating families for their time and contributions to this database, as well as the generosity of the donors who supported this program. We thank the staff at The Centre for Applied Genomics (TCAG) for support in data analysis. Funding was provided by the University of Toronto McLaughlin Centre and SickKids Foundation. S.W.S. holds the Northbridge Chair in Paediatric Research at the University of Toronto and The Hospital for Sick Children.
Author information
Authors and Affiliations
Contributions
M.S.R., N.B.S., J.L.H., N.H., E.S., T.S., M.M.A., B.Th., B.Tr., and S.W.S. were involved in the generation, analysis and interpretation of the genomic data. M.S.R., J.L.H., N.H., E.S., T.S., A.M.V., G.O., and C.M.F. reviewed the clinical data. M.S.R. and N.B.S. reviewed the literature for additional cases. M.S.R. wrote the manuscript. All authors provided feedback on the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.W.S. is the Editor-in-Chief for the journal npj Genomic Medicine, but was not involved in peer review process or decision making of the manuscript. At the time of this study and its publication, S.W.S. served on the Scientific Advisory Committee of Population Bio. Intellectual property from aspects of his research held at The Hospital for Sick Children are licensed to Athena Diagnostics and Population Bio. These relationships did not influence data interpretation or presentation during this study but are disclosed for potential future considerations. The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Reuter, M.S., Salazar, N.B., Howe, J.L. et al. UBR5 loss-of-function variants in autism spectrum disorder and intellectual disability: case series and review of the literature. npj Genom. Med. 11, 1 (2026). https://doi.org/10.1038/s41525-025-00536-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41525-025-00536-x