Abstract
Cultivation of microorganisms in space has enormous potential to enable in-situ resource utilization (ISRU) Here, we develop an autonomous payload with fully programmable serial passaging and sample preservation, termed the Modular Open Biological Platform (MOBP), and flight-test the MOBP aboard the International Space Station (ISS) by conducting enzymatic and microbial plastics upcycling experiments. The MOBP is a compact, modular bioreactor system that allows for sustained microbial growth via automated media transfers, such as those for sample collection and storage for terrestrial analyses, and precise data monitoring from integrated sensors. The MOBP was flight-tested with two experiments designed to evaluate biological upcycling of the plastic poly(ethylene terephthalate) (PET). The bioproduct βKA can be polymerized into a nylon-6,6 analog with improved properties for use in the production of a variety of materials. We posit the MOBP will aid in democratizing the execution of synthetic biology in spaceflight towards enabling ISRU.
Similar content being viewed by others
Introduction
Sustained human spaceflight is currently resource-limited by volume and weight restrictions. As we advance towards extended-duration spaceflight in low Earth orbit (LEO) and beyond, astronauts will be bound by the resources included in the initial launch payload. One attractive solution to this resource limitation is the development of technologies that can utilize local materials to generate new resources including water, construction materials, and propellant. Plastics are used ubiquitously in spaceflight for products ranging from food packaging, insulation, and machine parts. In-situ resource utilization (ISRU), the process of reusing and repurposing a wide range of materials, including plastics1,2,3, can enable longer and more adaptable manned space missions. ISRU of plastics commonly used in space applications, including poly(ethylene terephthalate) (PET)4,5, which has applications ranging from suit materials, parachutes, and building materials, has significant potential. Biologically-enabled ISRU can repurpose carbon into a diverse array of products and offers several advantages over traditional mechanical or chemical processes. For example, microorganisms can be easily transported, stored in a dormant state for long durations, engineered to biomanufacture a wide range of products (e.g., pharmaceuticals, polymers, waste processing), can perform in a range of environments, and the resulting bioprocesses can be low-energy and scale efficiently2. Recognition of these advantages has spawned the field of space bioprocess engineering, an emerging multidisciplinary field that aims to design, realize, and manage biologically-driven technologies with the goal of supporting life on extended space missions6,7,8. Several research groups have reported methods for cultivating and studying microbial growth in spaceflight9,10,11,12,13,14,15,16,17,18,19,20,21,22,23, largely focused on observing the impact of microgravity and spaceflight conditions on different species. However, biomanufacturing has yet to be fully implemented as a means of ISRU in space.
Biological approaches to plastics recycling have been developed in terrestrial environments but have yet to be applied for ISRU of plastics in space. Enzymes for the depolymerization of poly(ethylene terephthalate) to terephthalate (TPA) and ethylene glycol have been discovered and subsequently engineered24,25,26,27,28,29,30 enabling PET recycling via TPA repolymerization. Moving towards upcycling, or the conversion of waste plastics into products of higher value, microorganisms can be engineered to produce valuable chemicals from plastic feedstocks26,27,28, which is an advantage of biological approaches. For example, Pseudomonas putida KT2440 was engineered to convert TPA to β-ketoadipic acid (βKA), a precursor for performance-advantaged nylon-6,6, enabling upcycling of PET to a high-performance material30. Plastics recycling in LEO or beyond is gaining momentum in recent years, especially using 3D printing technologies13,14,15,31. The Refabricator is a notable example that has been installed on the ISS to demonstrate waste plastic recycling into high-quality 3D printing feedstocks31. However, to our knowledge, no existing approaches to plastic upcycling in space utilize biological or biochemical conversion processes, yet the application of biological ISRU for upcycling plastics in space has the potential to greatly impact manned spaceflight.
While biological ISRU has been proposed as a focus during space exploration, executing biological experiments during spaceflight is hindered by a variety of challenges6,32. First, the space, energy, and interaction time (e.g., astronaut hours) are highly restricted in spaceflight experiments, preventing the experimental supervision and real-time troubleshooting that is often required during biological experiments. Second, flight protocols and platform capacities differ drastically among mission profiles and launch partners, and launch opportunities are frequently delayed or scrubbed with little warning. Biological materials after revival are vulnerable to changes in time and environment, which jeopardizes the success of the experiments. The temperature variability in particular is a concern during the entire experimental timeline, as conditions are often uncontrolled during integration, transit, orbit, and return. Third, traditional culturing and sensing systems used in biology laboratories often require manual manipulation or are automated but highly specialized. While there are existing implementations of biological studies in spaceflight (see Table S1), most systems are standalone and often pre-designed with a fixed set of parameters, such as the total amount of media, collection samples, sensing capacity, hardware interfaces and cannot be easily adjusted or integrated with each other. The lack of versatile, cost-effective platforms with off-the-shelf components for space biology experiments is a significant obstacle for realization of research in this unique environment. Ultimately, these challenges impede the democratization of space biology and stymie the growth of the field. The development of enabling technologies, both hardware and wetware, along with comprehensive instructions to develop and use these systems, will be necessary for the growth of biologically-enabled ISRU in space and the accessibility of space-based biomanufacturing.
Here, we present a generalized payload, the Modular Open Biological Platform (MOBP), and a case study validating the MOBP for biological PET upcycling aboard the International Space Station (ISS). The MOBP is a compact, modular, and spaceflight-ready payload that enables automated cultivation of microorganisms via a programmable liquid transfer system. Key features of the MOBP are modular, open systems for employing “design-build-test-learn” (DBTL) processes, continuous microbial cultivation with sensing capacity (e.g., fully autonomous media transfers and preservation), and adaptable protocol execution (e.g., real-time response to mission profile changes). This platform addresses two of the main challenges faced by biological experimentation in space by functioning in an automated manner and being highly customizable. We validate the MOBP aboard the ISS for microbial PET upcycling and demonstrate conversion of TPA to βKA by MOBP-cultivated engineered Pseudomonas putida KT2440. In sum, we propose the MOBP as a platform for synthetic biology applications that will enable ISRU in space travel.
Results
Development of the MOBP hardware and software
The MOBP is an autonomous system for prototyping and evaluating bio-based ISRU in space. Its modular open system approach is critical for biology experiments with space-specific constraints, allowing for: (1) easy process integration and deinstallation; (2) independent storage and sterilization requirements for biological and non-biological materials; (3) high modularity to integrate new components and functionalities; and (4) rapid terrestrial prototyping under simulated space-like constraints. Most importantly, the modular open system enables a DBTLprocess, in which the biology experiment iterates together with flight hardware and software, and responds to unexpected constraints and changes (e.g., flight delays or mission profile changes). The total weight of the MOBP is 3.7 kg and its dimensions are 15.3 inches by 2.68 inches by 8.74 inches.
The system described in this manuscript was built for a biological plastics upcycling case study, one of many possible configurations and experimental goals. The experiment, termed MicroPET, was run in the MOBP and contained two independent experimental modules: an enzyme module (Module A) and microbial module (Module B) (Fig. 1). The subsequent sections describe the hardware and software used to implement the MOBP for MicroPET such that readers can adopt the system for their own experimental needs.
Module A executes enzymatic experiments. Chambers containing purified enzyme (Enzyme A1) and reaction buffer (Buffer A1) are pumped via Valve A1 and Pump A1 into Chamber A, containing the solid substrate, where the interfacial biocatalysis reaction occurs. At preset time-points, Valve A2 transfers liquid from Chamber A into Collection A, and the enzyme and buffer are replenished to initiate another round of biocatalysis. Module B is designed to implement experiments with microbial cultivation. Growth media (Media B2) is pumped via Pump B2 through the Revival Chip containing lyophilized bacteria and into Chamber B, the BioCell, to revive cells aboard the ISS. At set time-points, growth media from Media B1 is transferred into Chamber B via Pump B1 with excess liquid being transferred into Collection B1 via a commonly open valve (Valve B1). The media replenishing protocol above may be repeated at set time-points via a serial-connected valve array directing liquid transfer into Collection B2-B5, pre-filled with preservative (e.g., DNA/RNA shield for nucleic acid analysis), enabling up to five separate sample collections. Integrated temperature, humidity, and spectral sensors affixed above the Biocell (Chamber B) along with a microcontroller enable continuous data collection.
Liquid handling systems
The MOBP contains a modular liquid handling system that enables liquid transfers between various chambers at user-defined time-points or in response to sensor-based data prompts. The MOBP uses medical-grade bags (Table S2) for liquid containment to minimize the total weight and volume and enable modularity and robustness in the presence of external forces. Bioprocessing bags (Saint Gobain, FEP, Catalog #SGS05581, #SGS05108, #SGS05106) outfitted with Luer-lock connections were used for liquid storage of volumes from 3 to 30 mL. These bags can be modified by heat-sealing, allowing for accommodation of lower volumes and/or insertion of solid materials that cannot be added via the Luer-lock system. Bags were pre-sterilized and connected to other MOBP systems via a series of sterile Luer-locking parts.
A series of fixed-volume solenoid-type pumps (Lee company, LPM series) were utilized to move liquid between chambers. To accommodate the high pumping resolution required from Enzyme A1, the LPMA1250625L pump (dispensing volume ~25 μL) was selected. The fixed volume solenoid pumps can be programmed individually to provide accurate and repeatable dispense volumes in a small lightweight package. Pumps were flushed with 70% (v/v) ethanol to sterilize prior to each implementation. To enable multiple simultaneous liquid transfers (e.g., enzyme and buffer mixing in Module A) or liquid transfers to multiple collection bags with a single pump (e.g., Collection B1-B5 from Chamber B), several individual and a serial-connected array of solenoid valves (Lee Company, LHDA1233115H) were utilized.
The shared 12 V solenoid pump and valve electronic driving system allows a flexible, adaptable hardware design by simply swapping hardware and rerouting tubing connection. The system responds to a variety of different conditions in both spaceflight (e.g., mission profile changes, flight scrap, power outages) and microbial performance (e.g., change sampling time based on bacterial density, temperature, or humidity), and allows experiment iterations throughout the development. A manifest of all hardware used in the MOBP is provided in Table S2 and schematics are provided in the Supplemental Text.
Cell revival and cultivation
A key consideration for the MOBP was in-orbit cell revival as the time from payload assembly to experiment start-up on the ISS can exceed 14 days. To enable this, we designed a revival chip with a curved internal channel to allow maximum surface contact and mixing of revival media with lyophilized cell powder. The chip was 3D printed (BioMed Clear, Formlabs) and sterilized by ethylene oxide (EO) treatment. Lyophilized cell powder was loaded via the top port and capped with a rubber cap seal (Fig. 2A). Once in orbit and powered, 20 mL of growth media was pumped by a rotary diaphragm pump (KPV08, Koge Micro Tech Co.) through the revival chip into Chamber B to revive the cells.
For cell cultivation in Chamber B, we utilized the BioCell (BioServe Technologies), a well-established and space-tested cultivation system33 with transparent, gas-permeable film to enable aerobic cell growth (Fig. 2B). Notably, the BioCell can be utilized with a wide range of biological specimens beyond microorganisms, including mammalian cells and small eukaryotes, such that diverse research applications can be accessed in this system by altering the composition of wetware contained in the Media and Collection bags.
Temperature, pressure, humidity, and optical density sensors
To monitor bacterial growth in real-time, which can be quantified as a function of the optical density of the solution at 600 nm (OD600), we mounted a SMD Orange LED (HSMD-A1000, Broadcom) and a visible light sensor (AS7262, AMS) to directly face each other on opposing sides of the BioCell (Chamber B) (Fig. 2B). Temperature, humidity, and pressure sensors (BME280, BOSCH) were also mounted adjacent to the BioCell for data collection during the experiment. All sensor boards connect to the central microcontroller via two shared I2C bus, a commonly used signal protocol for easy compatibility and adaptation.
Sensor data collected during the flight are stored on an integrated SD card for data back-up. The system continuously outputs sensor data at 1-min intervals allowing direct monitoring of the experiment data when data-downlinking and ground station operation windows are available.
Control unit and software
The MOBP control unit is a Teensy 4.0 microcontroller pre-programmed to direct experiment protocols, integrate sensor data, and control fluid flow. The experiment protocol is pre-programmed in high-level human-readable functions (e.g., move liquid from Enzyme to Chamber A in Experiment 1) for users to edit easily without coding experience. The control unit reads from a coin battery-powered real-time clock (PCF8523, NXP) and handles the experiment protocol at 24 h intervals. This type of battery poses low risk for flight and has been approved for use in spaceflight within this context. Once the experiment starts, the microcontroller computes the time since the last execution based on the real-time clock reading. The cumulative time interval allows users to modify the experiments ranging from minutes to weeks in duration. The researcher can choose discrete time-points based on the specific research system and question.
In the event of a power loss during operation, we incorporated a safety mechanism to enable experimental continuation. This mechanism utilizes the real-time clock and data stored on the SD card to identify any temporal anomalies resulting from the power loss. Depending on the circumstances, this will activate one of two safety pathways. If power loss occurs without operation, the system logs the anomaly and proceeds to the next task in the original timeline. If the power loss takes place during operation, the system will abort the remaining operation, record the anomaly, and continue to the next task in the original timeline.
To evaluate and stress-test the liquid handling system, we developed a Python script (see Code Availability) that mirrors the code in the Teensy control unit to simulate the liquid volume changes in all bags. This enables the user to quickly and efficiently check the Teensy code to ensure the liquid transfers match their experimental goals.
MOBP flight test for proof-of-concept plastics upcycling aboard the ISS
To demonstrate the utility and space-test the MOBP, we designed two experiments to demonstrate plastic biological ISRU aboard the ISS. For the purpose of flight-testing Modules A and B independently, we designed an experiment where the two modules were not connected (e.g., no liquid transfers between the two) though future experimentation could easily integrate the two for more complex experimental designs by rerouting the flexible tubing and using existing hardware.
First, in Module A, enzymatic depolymerization of PET to TPA, enabling closed-loop PET recycling as TPA can subsequently be polymerized into PET. Second, in Module B, microbial upcycling of PET to βKA, demonstrating biological plastics upcycling as βKA can be polymerized into a nylon-6,6 analog. In Module B, the system was designed to enable sample preservation for post-flight analysis of nucleic acids (DNA and RNA) as well as proteins by providing collection bags that can be pre-filled with preservatives. In this way, the modularity of the MOBP enables a wide variety of other analyzes to be performed on a range of sample types, democratizing system biology of microbial growth in space.
Module A design and set-up
The PET-depolymerizing esterase PETase from Ideonella sakaiensis 206-F629 (IsPETase) was selected as it is well-studied and optimally active at 30 °C, the temperature at which the MOBP was anticipated to be incubated aboard the ISS. PETase catalyzes the depolymerization of amorphous PET to bis(2-hydroxyethyl)terephthalate (BHET), and further hydrolyzes BHET to mono(2-hydroxyethyl)terephthalate (MHET)29,34. MHET can spontaneously hydrolyze to TPA under low pH, high heat, or extended time periods; thus, by incubating PETase alone with amorphous PET, TPA can be generated35. Previous work has shown that the recovered TPA can be polymerized into PET with no loss of polymer properties24.
To determine PETase loading, we tested three concentrations each at a volume of 3 mL: 0.075, 0.15, and 0.30 mg/mL IsPETase with 250 mg of PET film. In a seven day reaction, 252, 292, and 88 μM TPA were detected from 0.075, 0.15, and 0.30 mg/mL enzyme loadings, respectively (Figure S1), indicating that 0.15 mg/mL was the optimal enzyme loading.
To load the MOBP, Enzyme A1 was pre-filled with 3 mL of 5.2 mg/mL IsPETase in 20 mM Tris at pH 8 and 300 mM NaCl. Buffer A1 was pre-filled with 23 mL of 100 mM NaCl and 50 mM Sodium phosphate pH 7.5 buffer, and Chamber A was pre-filled with a pre-weighed piece of amorphous PET film. A liquid transfer protocol was developed such that starting on Day 1 and proceeding every third day until Day 25, 0.075 mL from Enzyme A1 and 2.525 mL from Buffer A1 were pumped into Chamber A with overflow liquid being shunted to Collection A1 (Fig. 3). The accessible design of this platform allows for easy access to Chamber A (containing PET film) for post-flight analyses. The reaction products of PET degradation are stable at mesophilic temperatures, and thus can be pumped into Collection A1 without any storage buffer. However, this chamber can optionally be filled with a stabilizing storage buffer for accurate sampling of less stable reaction products. Two full biological replicates were prepared and loaded into the MOBP.
Module B design and set-up
A strain of P. putida KT2440 was previously engineered for conversion of TPA into βKA by (i) heterologously, constitutively, and chromosomally expressing the TPA transporter tpaK from Rhodococcus jostii RHA1, (ii) heterologously, constitutively, and chromosomally expressing the TPA catabolic operon tphA2IIA3IIBIIA1II from Comamonas sp. E6, and (iii) deleting the native 3-oxoadipate CoA transferase pcaIJ30. Genetic engineering for improved ethylene glycol and BHET/MHET utilization was also included, as ethylene glycol is another byproducts of PET deconstruction. The resulting genotype of strain AW165 is P. putida ΔhsdMR::Ptac:tphA2IIA3IIBIIA1II.E6 fpvA:Ptac:tpaKRHA1 Ptac:glcDEFG:PP_3749 ΔgclR::PETaseIs:MHETaseIs ΔpcaIJ.
Growth media was prepared as M9 minimal media supplemented with 0.06 M glucose to support cell growth, 0.01 M TPA for conversion to βKA, 25% (v/v) Percoll to minimize biofilm formation, and 50 μM ampicillin to minimize the risk of contamination as P. putida is naturally resistant to Ampicillin. To ensure growth in the BioCell system, strain AW165 was cultivated in growth media in either BioCells, Erlenmeyer flasks without agitation, or Erlenmeyer flasks with continuous agitation at 225 rpm. Growth was measured as OD600 over the course of five days. Growth was observed in all conditions; importantly, in the BioCell, glucose was utilized and TPA was converted to βKA (Table S3, Figure S2) demonstrating viability of the cultivation platform.
MOBP compartments were pre-filled as follows: 32 mL growth media in Media B1, 45 mL growth media in Media B2, 32 mL growth media in Chamber B, 10 mL RNA/DNA shield in Collection B2–B4, 10 mM DNA/RNA Shield in Collection B5, and lyophilized cells in the revival chip. A liquid transfer protocol was developed such that on Day 1, 10 mL of Media B1 was pumped through the Revival Chip and into Chamber B, and the cells were allowed to revive for five days before proceeding. Then, on Day 6 and every fourth day thereafter until Day 26, 5 mL from Media B1 was pumped into Chamber B, and the displaced volume was moved into Collection B2, B3, or B4, with the exception of Day 10 when an additional 10 mL was moved into Collection B5. This protocol was chosen to enable weekly sampling for nucleic acids (Collection B2–B4) and a single time-point for proteomics (Collection B5). Two full biological replicates were prepared and loaded into the MOBP (Fig. 2D). The full schedule of liquid transfers programmed to execute MicroPET within the MOBP is shown in Fig. 3.
Flight-test aboard SpaceX CRS-26
The MOBP components were prepared and integrated on Nov. 7–8, 2022 prior to hand-off to the integration partner NanoRacks on Nov. 9, 2022. SpaceX CRS-26, a commercial resupply service mission to the ISS, launched on Nov. 26, 2022 (delayed due to weather) from the Kennedy Space Center, returned to Earth via the Gulf of Mexico on Jan. 11, 2023, and was recovered by SpaceX’s recovery vessel Megan. The payload was received by the authors on Jan. 21, 2023. Thus, MOBP was on-Earth awaiting launch without power for 13 days, aboard the ISS for 43 days, and in transit after return for 10 days.
Data logs from the flight-test indicate the MOBP only had power for seven of the 30 operation-related days, from Days 1 to 4 and Days 8 to 10, due to a state-wide power interruption and malfunctions within the NanoRacks BlackBox system. Given this loss of power, only partial commands between Days 1 and 10 were executed, and none after Day 10. For Module A, the enzyme and buffer were moved into Chamber A on Day 1, and again on Day 10. For Module B, the initial cell revival protocol was executed on Day 1, and the transfer of media from Media B2 into Chamber B allowing liquid to pass into Collection B1 and B5 occurred on Day 10. Temperatures were higher and more variable than expected, ranging from 30–47 °C (Figure S3), likely due to irregular functioning of the circulation fans installed in the NanoRacks BlackBox system that were meant to cool the MicroPET experiment. Humidity generally tracked temperature, ranging from 68-100% across the experiment.
Upon receiving the MOBP, the physical condition was inspected and the volumes and OD600 of each compartment were measured (Fig. 4, Tables S4, 5, Figures S5–8). Module A components were visually undamaged for both replicates, though the final volume in Chamber A was below the expected value, likely due to the reduced number of liquid transfers. For Module B, there was a visible leak from Collection B5 in Replicate 1 and dark brown material visible in the tubing from Chamber B to the Collection bags (which we hypothesize is dried residue from the media and could be avoided by an additional tube locking mechanism), and final volumes were lower than expected. For the hardware, there was visible erosion in the metal valves and a deformation in the PVA printed box. Overall, there were volume losses that are difficult to account for due to the loss of power and therefore missing data logs. Nonetheless, due to the two liquid transfers that were executed during the flight test, analysis of chambers with collected samples was conducted.
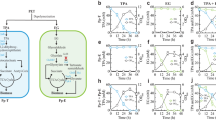
A Photographs of Chamber B (the BioCell) and Collection B1 and B5 (the two chambers that received liquid transfers during the flight test) post-flight for Experiment 1. Photographs of all chambers are provided in Figure S5, 6. B Growth of the engineered Pseudomonas putida KT2440 strain AW165 as measured by OD600. C Amount of βKA produced from TPA by cell cultivations. βKA abiotically decarboxylates to levulinic acid under high heat or low pH, so is quantified via levulinic acid, as described previously18.
Data logs indicate that liquid from Chamber B was transferred to Collection B1, which contained no preservative, and Collection B5, which contained DNA/RNA Shield, on Day 10. Visual observation and OD600 measurements corroborate the data log, with the OD600 in Collection B1 nearly matching that observed Chamber B for both replicates. The similar OD600 between the undiluted Collection bag and the BioCell also corroborate that no further liquid transfers were executed after Day 10 which would have diluted the BioCell. Further, both compartments exhibited similar OD600 measurements (Fig. 4B), suggesting stationary phase was reached and to a similar extent across the replicates.
βKA quantification analysis was conducted for all three chambers. βKA abiotically decarboxylates to levulinic acid under environmental conditions of high heat or low pH. No βKA was observed, but levulinic acid was observed, indicating that spontaneous decarboxylation occurred to completion (Fig. 4C). Thus, here we report βKA data from levulinic acid analysis, as has been previously described18. All three analyzed chambers had βKA, with levels in Chamber B and Collection B1 being higher than Collection B5, as would be expected as the latter was diluted with DNA/RNA Shield. βKA yields were not calculated as media volumes and thereby moles of TPA delivered were uncertain given the power losses. Nonetheless, the presence of levulinic acid in the microbial cultivation and sampling chambers indicates that AW165 was successfully revived and catalyzed the conversion of TPA to βKA while aboard the ISS.
Discussion
The main contributions of this work are two-fold: first, the development of a highly versatile and open biological cultivation system, the MOBP, for unmanned biological experimentation in spaceflight, and second, validation of the MOBP system for biological plastics upcycling by an engineered microorganism aboard the ISS. Overall, the system performed as designed during flight-testing, though sustained loss of power prevented the full realization of the experimental design. Still, the MOBP enabled microbial conversion of TPA, the monomer of PET, to βKA, the monomer of a performance-advantaged nylon-6,6 analog, demonstrating biological plastics upcycling aboard the ISS.
The MOBP was designed to enable those with limited hardware or spaceflight experimentation experience to design and execute unmanned biological experiments. All the parts used in the MOBP are off-the-shelf or 3D printed, the software was written for easy programming, the primary cultivation chamber is compatible with a variety of biological systems ranging from mammalian cells to bacteria, and the physical layout (e.g., size and number of compartments) is extremely flexible. The physical dimensions and data and power interfaces could be modified to enable compliance with alternative integration partners. Future expansions of the modules to include, e.g., DNA synthesis modules could enable enactment of the “design-build-test-learn” cycle during spaceflight. Together, we posit that the open and modular nature of the MOBP will work towards the democratization of access to biological research in space, currently a challenge for progression in the field.
Revival of biological systems aboard the ISS is a hurdle for researchers that do not have access to hands-on experimentation (i.e., no astronaut intervention to revive glycerol stocks). Here, we solved the problem with a custom 3D-printed revival chip (Fig. 2A), which proved effective at reviving lyophilized P. putida aboard the ISS after > 13 days of MOBP storage without power (Fig. 4). However, this aspect of the system design can be improved: specifically, once integrated into the full system, the revival chamber is subjected to humidity through the tubing and therefore may not remain dry after prolonged exposure. Further, loading the lyophilized powder is a tedious process as the entry point is small and located at a single point above the S-curves. Future system designs should be explored to address these issues by, for example, including check valves and a clamshell style enclosure that would prevent ambient moisture from contacting the powder and enable more efficient loading, respectively.
Another hardware improvement would be adaptation of the 3D-printed backing to withstand higher forces, such as the gravitational forces encountered during transit from the Kennedy Space Center to the ISS. It would also be advantageous to separate the electronics and the wetware within the system, or add additional levels of containment to prevent any liquid leakage damaging the electronic hardware.
The absence of TPA in Chamber A indicates that either (i) no enzyme was delivered to the reaction chamber, or (ii) the enzyme delivered was inactive. Given our successful terrestrial tests with the system and the rating of the pump, we are inclined to favor (ii) as the likely explanation. Sensor data indicates that aboard the ISS, temperatures were temporarily in excess of 40 °C (Figure S3), and the presence of only levulinic acid further suggests that high temperatures were encountered. Wild-type IsPETase is not a thermostable enzyme29,34, and therefore could have been inactivated in the storage buffer during transit from the Kennedy Space Center to the ISS (for which we do not have sensor data), or onboard the ISS. We did not use a high-temperature enzyme, such as leaf-branch compost cutinase (LCC)24, because we anticipated a reaction temperature aboard the ISS close to 30 °C. However, given the lessons learned here, future experiments would have increased likelihood of success by making the following modifications: (i) provide enzyme(s) as a lyophilized powder instead of in a liquid buffer to minimize heat transfer during transit to the ISS, (ii) utilize enzyme(s) that can withstand high temperatures during transit34, or (iii) utilize enzyme(s) that function at elevated temperatures. Indeed, IsPETase has been heavily engineered, and additional PET esterases have been discovered34, resulting in high thermostability, providing several possible biocatalysts that could meet these criteria for future experimentation.
The elevated, variable temperature aboard the ISS recorded by the payload is consistent with previous experiments within the NanoRacks BlackBox system36 and indicates that this is an expected feature of the experimental environment. In addition to using enzymes better suited to high temperatures, the payload could be designed to include heat sinks or fans. As mentioned earlier, the temperature variability inherent in spaceflight experimentation is a persistent challenge, and future inclusion of a cooling component in the MOBP may make the payload more robust but would require higher electricity demands that may not be accommodated by all integration platforms.
As space missions increase in duration, the need for resource sustainability becomes more important. One approach to enable resource sustainability is biological recycling or upcycling of common materials into higher value and versatile products. Here, glucose was provided as a carbon and energy source to support molar conversion of TPA to BKA; utilization of co-located or gaseous feedstocks would improve feasibility for bioconversion during spaceflight. Just as one could envision a bank of vials containing microbes engineered for producing a whole host of pharmaceuticals, one could envision a bank of vials containing microbes engineered for production of new materials. Additional examples of MOBP use cases for ISRU applications include the microbial treatment of human wastewaters, biological conversion of CO2 into valuable products, or evaluation of microbial bioleaching of critical materials and metals from ore. However, to achieve this vision, strains engineered and validated in terrestrial environments must be tested in spaceflight to benchmark strain performance and stability. The MOBP provides a platform for new-to-space researchers to test their biological systems under the complex conditions of spaceflight, thus addressing a critical gap towards biological ISRU in space.
Methods
Construction and assembly of the MOBP
The MOBP is a modular system that combines off-the-shelf, flight-proof hardware and customized components for maximum flexibility and off-the-shelf implementability. The assembly of the full MOBP includes three main components, where the separation of hardware (Module A and Module B) and wetware allows for easy integration and a low risk of contamination (Supplementary Text).
The payload enclosure was 3D printed using Polylactic Acid (PLA) with high-density infill for increased durability. Helicoil inserts and magnets were installed on the enclosure floor and along the enclosure wall for securing the hardware mounting brackets inside and closing the two-part payload. The rest mounting parts, BioCell holders and LED holders were 3D printed using selective laser sintering with Nylon for durability and flexibility. The top enclosure contained two L-shaped assembly duplicates of OTS pumps, valves, and a middle strip area for electronics. After pumps, valves, and manifolds were assembled, the tubing was routed for connection to minimize bending, following the diagram in Fig. 1 Module A. The bottom enclosure contains the two Module B BioCell sub-assembly and the printed circuit board holding twelve collection valves. Temperature, humidity, and pressure sensors (BME280, BOSCH) and visible light sensor (AS7262, AMS) were mounted on the bottom of the BioCell holder adjacent to the BioCell. The sensor boards I2C buses were connected using Teflon-coated wires and bolted on the BioCell holder. The orange LED board was glued to the inside of the LED holder and the LED holder was bolted to the nut inserts inside the BioCell holder. All electronic wires were routed to have slight extra length to minimize stress. We estimate the total cost of the assembled MOBP including hardware, electronics, and consumables to be $20,000-25,000; it is important to note that this cost estimate is based on the specific configuration of the MOBP used in our study and may vary depending on the requirements of different experiments.
The revival system and the BioCell required sterilization prior to installation. The revival chip was manufactured through stereolithography 3D printing using biocompatible resin (BioMed Clear, Formlabs). The chip was designed to have a curved internal channel that allows maximum surface contact with the cell powder as our revival pump pushes growth media through it. After the printing process, the revival chip was post-processed by washing in 75% ethyl alcohol and cured in the UV light for 15 min. Ethyl alcohol was injected through the internal channel using a syringe to prevent the remaining resin from blocking the internal structure of the revival chip. Once the chip was fully cured, we applied an elastic rubber lid to seal the open outlet where the lyophilized cell would be inserted. The chip was sterilized by EO sterilization and kept in a sterilized bag until the time of use. Hardware functional testing for electronics and code was conducted at the point before wetware installation.
Operating code of the MOBP
The payload control unit is a Teensy 4.0 microcontroller that can be pre-programmed to direct experiment protocols, integrate sensor data and control fluid flow with a real-time clock (PCF8523, NXP) to enable documentation of any power outages. The fluid system handles the routine liquid transfers to maintain the environment for cell growth, using a collection of fixed-volume solenoid pumps and valves that dispense liquid at a resolution of 25uL. Apart from BioCell in Module B, the system uses medical-grade bioprocessing bags for liquid containment to minimize the total weight and volume. The operating code for MOBP is written in Teensyduino and starts by including various libraries such as TimeLib, TimeAlarms, Wire, SPI, Adafruit_Sensor, Adafruit_BME280, Adafruit_AS726x, Adafruit_MCP23X17, and RTClib. These libraries are used to interface with different sensors and modules.
Next, there are function prototypes for 26 different experiment days, numbered from day_1 to day_26. These functions are used to execute specific experiments on each day of the payload’s operation. The experimentArray is an array of function pointers that stores the addresses of each experiment function. This allows the code to easily access and execute the desired experiment based on the current day. The code then defines various pin mappings and addresses for different components connected to the Teensyduino, such as the base board connectors and MCP23X17 GPIO expanders. Further in the code, there are implementations of functions for controlling motors, switching IO ports, moving liquids between different chambers, switching collection bags, taking sensor data, initializing the SD card, and saving data to files. The loop function of the code waits for a serial start command from the user to begin the payload’s experiments. Once the command is received, the code calls the day_1 function to start the first day’s experiment. After each experiment, the code sets the next alarm to schedule the next experiment based on the defined delay between experiments. There is also a function for recovering the system after an unscheduled power loss. This function checks if the system was interrupted during an experiment or if it was off for a longer time than the experiment interval. If it can resume the experiment, it sets the next alarm and continues from where it left off. The rest of the code includes functions for reading data from sensors, saving sensor data to a file, and initializing the SD card and experimental log file. Overall, the code provides the functionality for controlling and managing the MicroPET payload, executing different experiments, collecting sensor data, and saving data to files.
To evaluate and stress-test the liquid handling system, we developed a Python script that mirrors the code in the Teensy control unit and simulates the liquid volume changes in all bags. We designed the automation to enable three main tasks: (1) replenish enzyme and buffer into the enzymatic chamber (Chamber A), (2) replenish media into the microbial cultivation chamber (Chamber B), and (3) preserve cultivations at defined time-points. The researcher can choose these discrete time-points based on the specific research system and question. We have utilized the same flight-ready system for on-Earth experiments ranging from days to weeks in duration, to study, iterate, and optimize the experimental design. We also programmed the system with multiple responses in case of power loss, given the timestamp provided by the real-time clock. The final code for executing the payload is available at: https://github.com/mitmedialab/MicroPET/.
Cultivation media, bacterial strains, and lyophilization
Pseudomonas putida KT2440 (ATCC® 47054) was engineered as described previously in ref. 30 by chromosomal gene deletions, chromosomal gene overexpression, and heterologous chromosomal gene expression generating strain AW165 with the following genotype: P. putida ΔhsdMR::Ptac:tphA2IIA3IIBIIA1II.E6 fpvA:Ptac:tpaKRHA1 Ptac:glcDEFG:PP_3749 ΔgclR::PETaseIs:MHETaseIs ΔpcaIJ, named AW165. AW165 was stored in 20% (v/v) glycerol at −80 °C. For shake flask experiments, precultures were prepared by reviving a sample of the glycerol stock into Miller’s LB and cultivating overnight at 30 °C and 225 rpm. Shake flask cultivations were performed in M9 minimal medium (6.78 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 2 mM MgSO4, 100 μM CaCl2, and 18 μM FeSO4) with glucose, terephthalate, Kanamycin, and/or Percol® supplemented to the concentrations indicated for each individual experiment. Cell density was measured as absorbance at 600 nm. For experiments in the MOBP, AW165 was freeze-dried (lyophilized) by OPS Diagnostics, LLC (Lebanon, NJ, USA) according to their standard protocols, stored in sealed vials at room temperature until use, and prepared and cultivated in the MOBP as described in “Sterilization and loading the MOBP with wetware”. All chemicals used are listed in Table S6.
Enzyme expression and purification
BL21 (DE3) Escherichia coli (NEB) competent cells were transformed with a pET21b(+) plasmid encoding the wildtype IsPETase sequence with a C-terminal hexa-histidine epitope tag. The IsPETase sequence is provided in the source datafile previously described34. A single colony from transformation was inoculated into a starter culture of lysogeny broth (LB) media containing 100 µg/mL ampicillin and grown at 37 °C overnight. The starter culture was inoculated at a 100-fold dilution into a 2xYT medium containing 100 µg/mL Ampicillin and grown at 37 °C until the optical density measured at 600 nm reached 0.6-0.8. Protein expression was induced by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Cells were induced at 20 °C for 24 h, harvested by centrifugation, and stored at -80 °C until purification. Harvested cells were resuspended in a lysis buffer (20 mM Tris HCl, pH 8.0, 300 mM NaCl, 10 mM imidazole) with a final concentration of 0.25 mg/mL lysozyme, and 12.5 U/mL DNase I. Cells were lysed by sonication (50% power, 20 s ON, 20 s OFF for a total of 2 min 20 s ON). Lysate was clarified by centrifugation at 40,000 x g for 40 min at 4 °C. Clarified lysate was filtered through a 0.45 µm PVDF membrane, then applied to a 5 mL HisTrap HP Ni-NTA column (Cytiva) using an ÄKTA Pure chromatography system (Cytiva) and eluted using a buffer comprising 300 mM NaCl, 500 mM imidazole, 20 mM Tris HCl, pH 8.0. Resulting fractions containing the protein of interest were pooled and dialyzed at room temperature (25 °C) using a 3.5 kDa molecular weight exclusion membrane into 300 mM NaCl, 20 mM Tris, pH 8.0 buffer. After 20 h of buffer exchange, samples were centrifuged and evaluated by SDS-PAGE with Coomassie staining. Pooled samples were concentrated using 3.5 kDa molecular weight cut-off spin columns. Total protein was assessed by BCA assay and the sample was stored at 4 °C.
Experimentation with PETase
For all enzymatic experiments, purified IsPETase was suspended in the reaction buffer (100 mM NaCl and 50 mM Sodium phosphate, pH 7.5) and added to a washed, sterilized (in 70% ethanol), and pre-weighed PET coupon (cut from GoodFellow catalog number ES301445 amorphous PET film to a final mass of 200–400 mg) in a sterile Saint Gobain bag (see materials). PET film was sterilized by incubation in 70%, and dried in a sterile biosafety cabinet. The enzyme reaction was always performed at 30 °C in a traditional cell-culture incubator. Reaction times, volumes, and component concentrations varied as specified for each experiment.
Sterilization and loading the MOBP with wetware
Ethylene oxide (EO)-compatible hardware was sterilized by EO sterilization, including all tubing, Luer-lock accessories, rubber stoppers, loading spatula, and revival cell. Non-compatible hardware, namely the solenoid pumps and PET plastic coupons, were sterilized via washing with 70% (v/v) ethanol in water. Reaction and storage bags (Saint Gobain) Bags and Biocells came pre-sterilized by their respective vendors. Other reaction liquids (media, buffers) were sterilized with 0.22 um filtration. The lysis preservative reagent DNA/RNA shield (Zymo) was not sterilized. The revival chip was loaded with lyophilized power by opening the rubber stopper and using a sterilized straw to scoop the lyophilized cell powder and insert in the open outlet, then reassembled the rubber stopper. The revival pump tubing was sterilized by washing with 70% (v/v) ethanol, connected with the revival bag on its inlet, and sealed with the revival chip and BioCell on its outlet. Once assembled, the pump was slotted into the BioCell holder and pressed down by the BioCell. We created an assembly manual (Supplementary Information) with illustrated guidance.
Analysis of analytes
β-ketoadipic acid (βKA) was analyzed as levulinic acid. The hydrolysis of βKA was completed by high-temperature conditions during sample transit, so no additional hydrolysis was conducted before analysis37. Analysis was performed using an Agilent 1290 series ultra-high-performance liquid chromatography (UHPLC) system coupled with an Agilent 6470 A triple quadrupole mass spectrometer. The mass spectrometer utilizes dual Agilent jet stream electrospray ionization (ESI) and analysis was run in negative ion mode. Levulinic acid and the deuterated internal standard (levulinic-d5 acid) were optimized prior to analysis including optimization of the quantify and qualifying multiple reaction monitoring (MRM) transitions, fragmentor voltages and collision energies presented in Table S7. Chromatography was achieved using a Phenomenex Luna 2.5 μm, 2 × 100 mm C18(2)-HST column held at a temperature of 40 C, samples and standards were injected at a volume of 10 μL. The mobile phases used were (A) 0.1% formic acid in water and (B) 0.1% formic acid in methanol at a flow rate of 0.5 mL/min. The gradient was as follows: 0-1 min 100% (A), 7.67 min (A) 50% (B) 50%, 9.33 min (A) 30% B (70%), 10.67 min (A) 30% (B) 70%, returned to starting conditions at 10.68 min and held for a total run time of 13.00 min. The ESI source parameters were set to: capillary voltage 3 kV, nozzle voltage 2 kV, drying gas temperature 300 °C, drying gas flow 7 L/min, sheath gas temperature 350 °C, sheath gas flow 11 L/min, and the ESI nebulizer gas was set to 35 psi. Results were obtained from a seven-point calibration curve ranging from 0.1 – 5 µg/mL with a quadratic fit applied and an r2 coefficient of ≥0.995. A calibration verification standard was run every 10 – 15 samples to monitor for instrument and detector drift. To verify no unhydrolyzed βKA remained in samples, the levulinic acid MRM transitions were monitored at a retention time determined by a βKA standard. The absence of a response in this chromatographic region illustrated a complete hydrolysis to levulinic acid. MassHunter Quantitative Analysis (QQQ) software version 10.1 was used to complete the data analysis.
Data availability
All data is provided in the Supplementary Information or the repositories referenced in Code Availability.
Code availability
The operating code, 3D printing files, and electronic files for the MOBP are all available on GitHub. You can access the complete repository containing all these resources at: https://github.com/mitmedialab/MicroPET. This repository includes the operating code, 3D printing files, and electronic files necessary for the MOBP project.
Abbreviations
- ISRU:
-
in-situ resource utilization
- MOBP:
-
Modular Open Biological Platform
- ISS:
-
International Space Station
- PET:
-
poly(ethylene terephthalate)
- TPA:
-
terephthalate
- βKA:
-
β-ketoadipate
- LEO:
-
low-Earth orbit
- BHET:
-
bis(2-hydroxyethyl) terephthalate
- MHET:
-
mono(2-hydroxyethyl) terephthalate
References
Anand, M. et al. A brief review of chemical and mineralogical resources on the Moon and likely initial in situ resource utilization (ISRU) applications. Planet. Space Sci. 74, 42–48 (2012).
Montague, M. et al. The role of synthetic biology for in situ resource utilization (ISRU). Astrobiology 12, 1135–1142 (2012).
Olsson-Francis, K. & Cockell, C. S. Use of cyanobacteria for in-situ resource use in space applications. Planet. Space Sci. 58, 1279–1285 (2010).
Elena Charola, A., Madden, O., DePriest, P. T., Cobb, K. C. & Koestler, R. J. The Age of Plastic: Ingenuity and Responsibility: Proceedings of the 2012 MCI Symposium (Smithsonian Institution Scholarly Press, 2017).
Hedayati, R. & Stulova, V. 3D printing for space habitats: requirements, challenges, and recent advances. Aerospace 10, 653 (2023).
Berliner, A. J. et al. Space bioprocess engineering on the horizon. Commun. Eng. 1, 1–8 (2022).
Santomartino, R. et al. Toward sustainable space exploration: a roadmap for harnessing the power of microorganisms. Nat. Commun. 14, 1391 (2023).
Sanders, L. M. et al. Biological research and self-driving labs in deep space supported by artificial intelligence. Nat. Mach. Intell. 5, 208–219 (2023).
Coil, D. A. et al. Growth of 48 built environment bacterial isolates on board the International Space Station (ISS). PeerJ 4, e1842 (2016).
Klaus, D., Simske, S., Todd, P. & Stodieck, L. Investigation of space flight effects on Escherichia coli and a proposed model of underlying physical mechanisms. Microbiology 143, 449–455 (1997).
Brown, R. B., Klaus, D. & Todd, P. Effects of space flight, clinorotation, and centrifugation on the substrate utilization efficiency of E. coli. Microgravity Sci. Technol. 13, 24–29 (2002).
Crabbé, A. et al. Transcriptional and proteomic responses of Pseudomonas aeruginosa PAO1 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl. Environ. Microbiol. 77, 1221–1230 (2011).
Kim, W. et al. Effect of spaceflight on Pseudomonas aeruginosa final cell density is modulated by nutrient and oxygen availability. BMC Microbiol 13, 241 (2013).
Kim, W. et al. Spaceflight promotes biofilm formation by Pseudomonas aeruginosa. PLoS ONE 8, e62437 (2013).
McLean, R. J., Cassanto, J. M., Barnes, M. B. & Koo, J. H. Bacterial biofilm formation under microgravity conditions. FEMS Microbiol. Lett. 195, 115–119 (2001).
Benoit, M. R. et al. Microbial antibiotic production aboard the International Space Station. Appl. Microbiol. Biotechnol. 70, 403–411 (2006).
Juergensmeyer, M. A., Juergensmeyer, E. A. & Guikema, J. A. Long-term exposure to spaceflight conditions affects bacterial response to antibiotics. Microgravity Sci. Technol. 12, 41–47 (1999).
Lam, K. S. et al. The effect of space flight on the production of actinomycin D by Streptomyces plicatus. J. Ind. Microbiol. Biotechnol. 29, 299–302 (2002).
Klaus, D. M. & Howard, H. N. Antibiotic efficacy and microbial virulence during space flight. Trends Biotechnol. 24, 131–136 (2006).
Nickerson, C. A. et al. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 68, 3147–3152 (2000).
Hammond, T. G. et al. Effects of microgravity on the virulence of Listeria monocytogenes, Enterococcus faecalis, Candida albicans, and methicillin-resistant Staphylococcus aureus. Astrobiology 13, 1081–1090 (2013).
Leys, N. M. E. J., Hendrickx, L., De Boever, P., Baatout, S. & Mergeay, M. Space flight effects on bacterial physiology. J. Biol. Regul. Homeost. Agents 18, 193–199 (2004).
Horneck, G. Survival of microorganisms in space: a review. Adv. Space Res. 1, 39–48 (1981).
Tournier, V. et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 580, 216–219 (2020).
Bell, E. L. et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 5, 673–681 (2022).
Kim, H. T. et al. Biological valorization of poly(ethylene terephthalate) monomers for upcycling waste PET. ACS Sustain. Chem. Eng. 7, 19396–19406 (2019).
Sullivan, K. P. et al. Mixed plastics waste valorization through tandem chemical oxidation and biological funneling. Science 378, 207–211 (2022).
Tiso, T. et al. Towards bio-upcycling of polyethylene terephthalate. Metab. Eng. 66, 167–178 (2021).
Yoshida, S. et al. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199 (2016).
Werner, A. Z. et al. Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab. Eng. 67, 250–261 (2021).
Prater, T. J. et al. NASA’s in-space manufacturing project: update on manufacturing technologies and materials to enable more sustainable and safer exploration. 70th International Astronautical Congress 2019, Washington, DC, 2019.
Afshinnekoo, E. et al. Fundamental biological features of spaceflight: advancing the field to enable deep-space exploration. Cell 183, 1162–1184 (2020).
BioCell/PHAB. BioServe Space Technologies. https://www.colorado.edu/center/bioserve/spaceflight-hardware/biocellphab (University of Colorado Boulder, 2018).
Erickson, E. et al. Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity. Nat. Commun. 13, 7850 (2022).
Brizendine, R. K. et al. Particle size reduction of poly(ethylene terephthalate) increases the rate of enzymatic depolymerization but does not increase the overall conversion extent. ACS Sustain. Chem. Eng. 10, 9131–9140 (2022).
Sharma, S. et al. Designing the organism-environment relationship (Massachusetts Institute of Technology, 2020).
Rorrer, N. A. et al. Production of β-ketoadipic acid from glucose in Pseudomonas putida KT2440 for use in performance-advantaged nylons. Cell Rep. Phys. Sci. 3, 100840 (2022).
Acknowledgements
The authors X.L., P.P., B.F., S.S., N.P.G., B.T.T., and A.E. thank the Office of Naval Research and Seed Health for funding this project, and S.A. for mission integration assistance. Funding for A.Z.W., E.E., K.J.R., M.A.I., N.P.M., and G.T.B. was provided by the U.S. Department of Energy, Office of Energy Efficiency and Renewable Energy, Advanced Materials and Manufacturing Technologies Office (AMMTO) and Bioenergy Technologies Office (BETO) and was performed as part of the BOTTLE™ Consortium and was supported by AMMTO and BETO under contract no. DE-AC36-08GO28308 with the National Renewable Energy Laboratory, operated by Alliance for Sustainable Energy, LLC. This material is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program under Award Number DE-SC0022024.
Author information
Authors and Affiliations
Contributions
X.L., P.P., C.E.M., A.Z.W. and E.E. conceptualized the project. X.L., P.P. and P.C. designed, built, and programmed the payloads. X.L., P.P., B.F., A.Z.W., E.E., N.P.M., P.C., K.A.R. and B.T.T. performed the experiments. X.L., P.P., B.F., S.S. and A.Z.W. integrated the payload. K.J.R. and M.A.I. performed the analytics. X.L., P.P. and A.Z.W. analyzed and visualized the data. X.L., P.P., B.F., A.Z.W., N.P.G., S.S. and B.T.T. wrote the manuscript. GTB, CEM, and AE provided resources and funding. All authors read, edited, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.Z.W. and G.T.B. have filed a patent on the strain utilized in this work (US Patent App. 17/198,230, 2021).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, X., Pataranutaporn, P., Fram, B. et al. Development and flight-testing of modular autonomous cultivation systems for biological plastics upcycling aboard the ISS. npj Microgravity 11, 23 (2025). https://doi.org/10.1038/s41526-025-00463-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41526-025-00463-2