Abstract
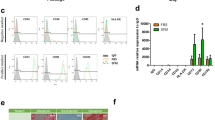
Despite its significant potential, cultivated fat manufacture remains a relatively inefficient process, as it primarily uses source preparations of animal-derived mesenchymal stem/stromal cells (MSC) that are poorly defined and highly heterogenous in nature, containing only relatively minor fractions of bona-fide adipocyte progenitors. The aim of this study was to use RNA-sequencing of clonal MSC populations from cattle to identify cell surface marker(s) of bona-fide pre-adipocytes to be used for efficient enrichment of MSCs, with a view to future cultivated fat applications. Adipose-derived MSC populations (n = 5 animals) were grown clonally from single cells and subsequently tested for adipogenic capacity. Adipogenic (A) and non-adipogenic (N) clones (n = 10/group) thus identified were bulk RNA-sequenced. A total of 35 cluster of differentiation (CD) genes were identified among differentially expressed transcripts, of which CD13, CD141, CD36, CD55 and CD34 were selected for further testing using flow cytometry in bovine MSCs. All antigens except CD13 were detected at negligible levels. FACS was then used to sort MSCs (n = 4 animals) into CD13+ and CD13- fractions. Sorted CD13+ cells were larger and flatter, grew significantly slower, and expressed substantially higher levels of adipogenic regulators (PPARG and CEBPA) compared to CD13- cells. Moreover, on average, adipogenic efficiency was 10.3-fold higher in CD13+ than CD13- cells, as demonstrated by BODIPY staining and confirmed by differential expression of mature adipocyte markers (FABP4, ADIPOQ, LEP), while expression of alternative lineage markers (chondrogenic and osteogenic) and ability to differentiate into bone and cartilage were both similar for CD13+ and CD13- cells. In summary, we identified CD13 as a bona-fide marker of pre-adipocytes in bovine and demonstrated the potential of using CD13-based cell selection for enriching MSC populations for cultivated fat purposes.
Similar content being viewed by others
Data availability
Raw fastq files and counts from RNA sequencing were deposited in NCBI’s GEO under Series GSE281680. All other data is available within the manuscript.
References
Yuen et al. Perspectives on scaling production of adipose tissue for food applications. Biomaterials 280, 121273 (2022).
Ferrero, R., Rainer, P. & Deplancke, B. Toward a consensus view of mammalian adipocyte stem and progenitor cell heterogeneity. Trends Cell Biol. 30, 937–950 (2020).
Adler, C. E. & Sanchez Alvarado, A. Types or states? Cellular dynamics and regenerative potential. Trends Cell Biol. 25, 687–696 (2015).
Crisan, M. et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3, 301–313 (2008).
Palani, N. P. et al. Adipogenic and SWAT cells separate from a common progenitor in human brown and white adipose depots. Nat. Metab. 5, 996–1013 (2023).
Dohmen, R. G. J. et al. Muscle-derived fibro-adipogenic progenitor cells for production of cultured bovine adipose tissue. npj Sci. Food 6, 6 (2022).
Ishida, Y., Mabuchi, Y., Naraoka, Y., Hisamatsu, D. & Akazawa, C. Conservation of markers and stemness in adipose stem and progenitor cells between cattle and other species. Int J. Mol. Sci. 24, 11908 (2023).
Song, W.-J. et al. Identification of porcine adipose progenitor cells by fluorescence-activated cell sorting for the preparation of cultured fat by 3D bioprinting. Food Res. Int. 162, 111952 (2022).
Heyman, E. et al. Validation of multiparametric panels for bovine mesenchymal stromal cell phenotyping. Cytom. A 103, 744–755 (2023).
Danev, N., Li, G., Duan, J. E. & Van de Walle, G. R. Comparative transcriptomic analysis of bovine mesenchymal stromal cells reveals tissue-source and species-specific differences. iScience 27, 108886 (2024).
Gao, H. et al. CD36 is a marker of human adipocyte progenitors with pronounced adipogenic and triglyceride accumulation potential. Stem Cells 35, 1799–1814 (2017).
Russell, K. C. et al. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells 28, 788–798 (2010).
Rostovskaya, M. et al. Clonal analysis delineates transcriptional programs of osteogenic and adipogenic lineages of adult mouse skeletal progenitors. Stem Cell Rep. 11, 212–227 (2018).
Cawthorn, W. P., Scheller, E. L. & MacDougald, O. A. Adipose tissue stem cells meet preadipocyte commitment: going back to the future. J. Lipid Res. 53, 227–246 (2012).
Mitchell, J. B. et al. Immunophenotype of human adipose-derived cells: temporal changes in stromal-associated and stem cell-associated markers. Stem Cells 24, 376–385 (2006).
Esteves, C. L. et al. Isolation and characterization of equine native MSC populations. Stem Cell Res. Ther. 8, 80 (2017).
Wang, S. et al. Effects of long-term culture on the biological characteristics and RNA profiles of human bone-marrow-derived mesenchymal stem cells. Mol. Ther. Nucleic Acids 26, 557–574 (2021).
Maumus, M. et al. Native human adipose stromal cells: localization, morphology and phenotype. Int. J. Obes. 35, 1141–1153 (2011).
Mina-Osorio, P. The moonlighting enzyme CD13: old and new functions to target. Trends Mol. Med. 14, 361–371 (2008).
Muñiz, C. et al. Ex vivo identification and characterization of a population of CD13(high) CD105(+) CD45(-) mesenchymal stem cells in human bone marrow. Stem Cell Res. Ther. 6, 169 (2015).
Yue, Y., Zhang, L., Zhang, X., Li, X. & Yu, H. De novo lipogenesis and desaturation of fatty acids during adipogenesis in bovine adipose-derived mesenchymal stem cells. Vitr. Cell Dev. Biol. Anim. 54, 23–31 (2018).
Rahman, M. M. et al. CD13 promotes mesenchymal stem cell-mediated regeneration of ischemic muscle. Front. Physiol. 4, 402 (2014).
Iaffaldano, L. et al. High aminopeptidase N/CD13 levels characterize human amniotic mesenchymal stem cells and drive their increased adipogenic potential in obese women. Stem Cells Dev. 22, 2287–2297 (2013).
Boyle, K. E. et al. Mesenchymal stem cells from infants born to obese mothers exhibit greater potential for adipogenesis: the healthy start babyBUMP project. Diabetes 65, 647–659 (2016).
Grandl, G. et al. Depot specific differences in the adipogenic potential of precursors are mediated by collagenous extracellular matrix and Flotillin 2 dependent signaling. Mol. Metab. 5, 937–947 (2016).
Michelotti, T. C. et al. Single-nuclei analysis reveals depot-specific transcriptional heterogeneity and depot-specific cell types in adipose tissue of dairy cows. Front. Cell Dev. Biol. 10, 1025240 (2022).
Mitić, R., Cantoni, F., Börlin, C. S., Post, M. J. & Jackisch, L. A simplified and defined serum-free medium for cultivating fat across species. iScience 26, 105822 (2023).
Thrower, T., Riley, S. E., Lee, S., Esteves, C. L. & Xavier Donadeu, F. A unique spontaneously immortalised cell line from pig with enhanced adipogenic capacity. npj Sci. Food 9, 52 (2025).
Kamaldinov, T., Erndt-Marino, J., Levin, M., Kaplan, D. L. & Hahn, M. S. Assessment of enrichment of human mesenchymal stem cells based on plasma and mitochondrial membrane potentials. Bioelectricity 2, 21–32 (2020).
van Velthoven, C. T. J., de Morree, A., Egner, I. M., Brett, J. O. & Rando, T. A. Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 21, 1994–2004 (2017).
Dan-Jumbo, S. O. et al. Derivation and long-term maintenance of porcine skeletal muscle progenitor cells. Sci. Rep. 14, 9370 (2024).
Merrick, D. et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 365, 2501 (2019).
Weatherall, E. L. et al. Differentiation potential of mesenchymal stem/stromal cells is altered by intrauterine growth restriction. Front Vet. Sci. 7, 558905 (2020).
Acknowledgements
The authors are grateful to the Roslin Institute’s Bioimaging Facility staff for support with microscopy and cell sorting, and to Isabella Donadeu and James Nathan for assistance with image analyses and qPCR analyses, respectively. This study was funded by an Innovate UK SMART award (project 10031578) to Roslin Technologies Ltd. and F.X.D, an Impact Acceleration Account award to F.X.D (IRM Ref: RE17173), and an Institute Strategic Programme Grant to The Roslin Institute. We acknowledge Roslin Technologies for their collaboration on this study and thank Dr. Madeleine Carter, Dr Deepika Rajesh and Dr. Joe Mee for their contributions.
Author information
Authors and Affiliations
Contributions
F.X.D. conceived the study, S.L, T.T, and S.E.R. generated and analysed experimental data, S.L., T.T., C.L.E., and F.X.D. interpreted the data and wrote the manuscript, and all authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
FXD declares that the project was funded by a joint award with Roslin Technologies Ltd. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, S., Thrower, T., Riley, S.E. et al. CD13 is a bona-fide marker of bovine pre-adipocytes with potential in cultivated fat applications. npj Sci Food (2026). https://doi.org/10.1038/s41538-026-00711-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41538-026-00711-z