Abstract
T cells mediate pathogenesis of several autoimmune disorders by recognizing self-epitopes presented on Major Histocompatibility Complex (MHC) or Human Leukocyte Antigen (HLA) complex. The majority of autoantigens presented to T cells in various autoimmune disorders are not known, which has impeded autoantigen identification. Recent advances in immunopeptidomics have started to unravel the repertoire of antigenic epitopes presented on MHC. In several autoimmune diseases, immunopeptidomics has led to the identification of novel autoantigens and has enhanced our understanding of the mechanisms behind autoimmunity. Especially, immunopeptidomics has provided key evidence to explain the genetic risk posed by HLA alleles. In this review, we shed light on how immunopeptidomics can be leveraged to discover potential autoantigens. We highlight the application of immunopeptidomics in Type 1 Diabetes (T1D), Systemic Lupus Erythematosus (SLE), and Rheumatoid Arthritis (RA). Finally, we highlight the practical considerations of implementing immunopeptidomics successfully and the technical challenges that need to be addressed. Overall, this review will provide an important context for using immunopeptidomics for understanding autoimmunity.
Similar content being viewed by others
Introduction
T cells form a highly antigen-specific arm of the adaptive immune system1. T cells achieve this specificity through their surface T Cell Receptors (TCRs), which are diversified through V(D)J recombination to generate a large number of unique clones2,3. T cells can either have TCRalpha and beta chains (αβT cells), or TCRgamma and delta chains (γδT cells). Classically, αβT cells recognize peptide epitopes presented on MHC/HLA, whereas γδT cells recognize non-peptide ligands on non-classical MHC such as CD1d and MR14,5. While presentation through and recognition of non-classical MHC is important, here, we will focus on presentation of peptide epitopes to T cells by classical MHC. For the purposes of this review, we will use MHC and HLA interchangeably, with HLA as the preferred terminology for human alleles. There are two distinct classes of T cells, CD8+ T cells recognize epitopes on class I MHC, whereas CD4+ T cells recognize epitopes on class II MHC. The epitopes presented on class I MHC are typically 8-12 amino acids in length, whereas those presented by class II MHC can range from 10-25 amino acids6,7. Class I MHC are found on all nucleated cells; whereas class II MHC are found typically on professional antigen presenting cells (APCs) such as B cells, Dendritic Cells, and Macrophages8,9. Central and peripheral tolerance mechanisms typically restrict self-reactive T cells. T cells that recognize self-antigens in MHC molecules can be unleashed either as a failure of tolerance and/or therapeutic blockade of immune checkpoints10,11. The recognition of self epitopes presented on MHC is critical for developing autoimmunity.
The pathways and sources of epitopes presented on these molecules are distinct, and together extensively sample the extracellular and intracellular proteome12,13,14. The MHC Class I pathway presents peptides from intracellular sources, such as viral or endogenous proteins, to CD8+ T cells14. These proteins are degraded by the proteasome, and the resulting peptides are transported into the ER by TAP, where they are loaded onto class I MHC molecules. In contrast, the class II MHC pathway captures extracellular proteins, which are internalized, processed in endosomes, and presented on class II MHC molecules to CD4+ T cells. Cross-presentation involves the uptake of extracellular antigens by dendritic cells, which then process these antigens via either a vacuolar pathway or a cytosolic pathway. In the vacuolar pathway, antigens are degraded within endosomal compartments and loaded onto class I MHC molecules. In the cytosolic pathway, Dendritic cells can translocate the extracellular antigens into the cytosol, where they are processed by the proteasome. This mechanism allows Dendritic cells to activate CD8+ T cells and initiate cytotoxic responses against pathogens or tumors that do not directly infect them. The ability of MHC molecules to process and present epitopes from self and foreign proteins is critical for adaptive immunity15,16,17. Another important property of MHC is its high degree of polymorphism, with thousands of class I (HLA-A, HLA-B, HLA-C) and class II (HLA-DP, HLA-DQ, HLA-DR) alleles. Even inbred mouse strains have a diverse set of MHC (class I alleles H2-K, H2-D, H2-L and class II alleles H2-A and H2-E). This serves to diversify the T cell response to pathogens at a population level but adds complexity to studying the repertoire of peptides presented by them18,19,20.
Association of HLA alleles with autoimmunity
The HLA locus is one of the highest disease-associated genetic loci in autoimmunity21,22. Genetic variation in HLA alleles can confer protection from disease, e.g., HLA-DR2 in T1D23 or can contribute to the risk of developing autoimmunity, e.g., HLA-DQ2, DQ8, DR3, DR4, and HLA-A*02:01 in Type 1 Diabetes24,25. Similarly, in RA, HLA-DRB1*04:01, 04:04, 04:05, 01:01, and 10:01 are risk alleles26,27. The same HLA allele can be both protective and pathogenic for different autoimmune diseases. One of the most well-studied examples of such alleles is HLA-DR15 (HLA-DRB1*15:01), which confers heightened risk of developing Multiple Sclerosis, but is protective in T1D28,29,30,31,32.
Variations in HLA alleles can predispose to or confer protection from autoimmunity by influencing the repertoire of peptides presented on them, and thereby shaping the adaptive immune response. Different HLA alleles have distinct peptide-binding preferences, resulting in unique peptide repertoires presented on the cell surface33,34,35. A striking example of how HLA-specific peptide preferences can positively and negatively influence immunity is found in HLA-B*27:05, a class I HLA allele. HLA-B*27:05 is well known to be enriched in individuals who can naturally control HIV infection, possibly by limiting viral escape from T cells36,37,38,39. On the other hand, HLA-B*27:05 is also strongly associated with Ankolysing Spondylitis40, by possibly presenting self-peptides that may mimic microbial peptides. Another key set of studies providing a mechanistic explanation of how minute variations in HLA can affect immune repertoires comes from T1D. The accepted animal model of T1D is Non-Obese Diabetic (NOD) mice, which bear the class II MHC allele, I-Ag7. I-Ag7 possesses similar binding properties to the human T1D risk alleles, HLA-DQ8 and DQ241. Studies have shown that these alleles have a neutral residue at the 57th position of the MHC-II β chain42, resulting in a unique peptide-binding pocket. This limits the diversity of islet associated self-peptide repertoire, thereby restricting the islet-reactive TCR repertoire43,44. Upon modification of I-Ag7 to change the β57 residue to D/E, the TCR repertoire to insulin was altered, suggesting that changes at a single position in HLA/MHC can have profound effects on autoimmunity. These studies underscore the importance of identifying the peptides presented by disease-associated MHC/HLA to gain a better understanding of autoimmunity. An extensive literature and public database search was done to highlight some key HLA associations, epitopes, and antigens in context to autoimmune diseases which are listed in Table 1.
Immunopeptidomics: The marauders’ map of MHC-presented epitopes
The importance of MHC and epitope presentation has been documented since the mid-20th century. Seminal work on tissue rejection and transplantation immunology in the early to mid-20th century, led to the discovery of the MHC genetic locus, first in mice (H-2)45 and then in HLA locus in humans46. In the 1960s and 1970s, the major focus was on describing the genetics, structure, and function of MHC, revealing their critical role in antigen presentation to T cells47,48,49,50,51,52,53. Subsequent studies throughout the latter half of the 20th century revealed the finer details of T cell recognition of class I and class II MHC, establishing the principle of MHC as peptide receptors that present degraded proteins1. The molecular basis of MHC restriction was firmly established by elucidating the crystal structure of HLA-A254, and nearly a decade later, in 1996, the TCR-peptide-MHC complex55. Our understanding of the landscape of epitopes presented by MHC has evolved alongside these discoveries. In the past decade, interest in identifying the peptides bound to MHC, collectively referred to as the ‘immunopeptidome’, has exploded across the biomedical research community56,57. This has been enabled by the yet nascent field of ‘immunopeptidomics’, which lies at the intersection of immunology and proteomics, and uses high-resolution mass spectrometry (MS) to identify and quantify the peptide repertoire presented by MHC58. Since its inception, immunopeptidomics has facilitated our understanding of T cell responses, and has greatly enhanced the identification and profiling of antigen-specific T cells59. Here, we will review how immunopeptidomics has been deployed to understand key autoantigens in autoimmune disorders. First, we will lay out the fundamentals and the technical considerations of implementing immunopeptidomics. Second, we will discuss how immunopeptidomics has transformed our understanding of post-translationally modified (PTM) epitopes. Third, we will discuss how immunopeptidomics has facilitated the identification of autoantigens and autoreactive T cells in three exemplary autoimmune disorders. Finally, we will summarize the outstanding challenges and provide future perspectives on utilizing immunopeptidomics for autoantigen discovery.
Immunopeptidomics: fundamentals and practical considerations
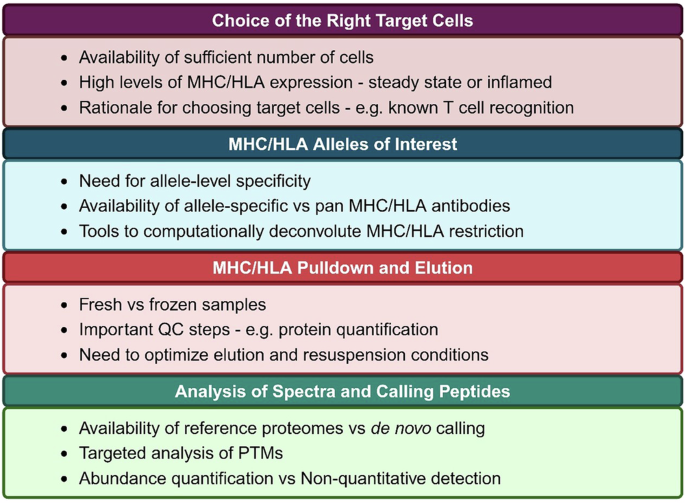
Immunopeptidomics relies on MS for identification of protein fragments, similar to proteomics. However, given that peptide-MHC is a binary complex that is non covalently bound, peptides must be decoupled from MHC prior to MS. The four major steps in immunopeptidomics are: 1) Pulldown of MHC complexes using MHC-specific antibodies immobilized on beads; 2) Elution of peptides off of MHC using non-enzymatic mild acid treatment; 3) Peptide purification with either C18 reverse-phase separation or size-based methods such as size exclusion chromatography or filtering through a specific molecular weight cut off filter. Purified peptides are then subjected to MS; and 4) Bioinformatic identification of peptides from MS spectra. Each of these steps needs to be carefully designed, with several experiment-specific considerations. These are highlighted in Fig. 1. Some of the major considerations are: the need for a large number of starting cell numbers or amount of tissue, levels of MHC/HLA expression on the target cells, and the availability of antibodies specific to MHC/HLA alleles under investigation. The pioneering study that developed immunopeptidomics60 reported only tens of HLA-bound peptides in a single analysis from billions of cells. Recent advances in MS methods have enabled detection of peptides from limited sample with sensitivity, thus allowing for detection of antigens from primary cells and rare cell polulations61. At present day, samples less than a billion cells can yield tens of thousands of peptides, including those with PTMs62. The technical advances are largely fueled by Liquid Chromatography and tandem MS (LC-MS/MS), as well as sophisticated computational tools to identify and quantify spectra. For instance, peptide purification techniques have evolved to minimize peptide loss. They can range from using a molecular weight cut off filters or size exclusion chromatography to more sophisticated C18 reverse-phase chromatography that can separate compounds based on hydrophobicity. Advances in sample preparation during MS have also led to increased yields of peptides. For instance, a recent report used acetonitrile fractionation followed by introduction of ion mobility during gas phase separation to increase the number of detected peptides from the same samples by 2-5 fold63. Similarly, Gravel et al., developed ion mobility separation-based time-of-flight (TOFIMS) MS to increase the sensitivity of immunopeptidomics64. Several computational tools have led to increased sensitivity of detection and wide accessibility to users. For instance, ‘Immunolyser’ is a web-based tool that allows a standardized and streamlined workflow for immunopeptidomics that is accessible to researchers without any prior experience in MS65. Similarly, SysteMHC Atlas v2.0 is a resource that has collected over 1 million peptides across over 7000 MS studies, and developed a suite of computational tools for analyses of PTMs, which has led to identification of over 470,000 modified peptides66. A key consideration for calling peptides is also the selection of the appropriate databases. For instance, using the annotated genome will miss a large number of peptides that may be derived from unannotated ORFs67,68. Moreover, implementing machine learning algorithms such as PROSIT, and MS2rescore, MSbooster have led to increased sensitivity and reduced false negative rates69,70,71,72. Finally, while all these advances have enhanced peptide detection individually, combining them in various ways has synergistically led to significantly better outputs from immunopeptidomics73.
Post-translationally modified epitopes
PTMs refer to chemical modifications of amino acid side chains occurring after their translation74. PTMs can profoundly impact the structure, function, and localization of proteins75. Of the 400 different types of PTMs that have been described in humans, phosphorylation, acetylation, and ubiquitination occur most frequently and are the most-well studied76,77,78,79. PTMs also alter the immunogenicity of peptide epitopes, which can be important in autoimmune disorders such as T1D and RA. The first demonstration of a PTM epitope presented on MHC was in melanoma, where a Tyrosinase derived epitope was found to be deamidated80. Since then, numerous studies have profiled PTM epitopes on MHC81,82,83,84,85,86,87,88. It is estimated that peptides containing PTMs make up ~10% of the human immunopeptidome89,90,91,92. Importantly, dysregulation of PTMs is increasingly being implicated in the pathogenesis of autoimmune disease93,94. The contribution of PTMs to autoimmunity is manifold, involving a combination of host genetic factors and environmental exposures. Mechanistically, PTM of self-proteins can generate new epitopes, known as neoepitopes, capable of eliciting robust immune responses and breaking immune tolerance95. PTM epitopes can alter binding to MHC and/or TCRs, thereby creating immunogenic neoepitopes96,97,98. PTMs add a layer of complexity to immunopeptidomics studies because of their low abundance, altered spectral profiles, and computational hurdles in identification59,92. PTMs may also be detected as an artifact of ionization that occurs during MS. It is appreciated that the immunopeptidomes of class I and II MHC differ in the types, positions, and ratios of PTMs99. These preferences likely reflect: a) distinct research questions and model systems used, b) inherent differences in antigen processing and presentation between class I and II MHC, and c) diversity of pathways leading to PTMs89,99. While PTMs are often a small fraction of the immunopeptidome, their importance as autoantigens is outsized. For instance, a major epitope known as 2.5HIP (a hybrid peptide formed by post-translational splicing of Insulin and Chromogranin), was shown to be an essential antigen in NOD mice. However, it is often not detected in immunopeptidomics datasets. The importance of PTMs may often be disease-specific, and therefore PTM identification might not be relevant in all cases.
How can immunopeptidomics fuel antigen discovery?
Recent advances in single cell TCR sequencing have led to a large number of disease-associated T cells being profiled, however, the knowledge of their cognate epitopes is lagging behind by orders of magnitude100. Experimentally, there are two types of approaches used for T cell epitope discovery. Antigen-directed approaches start with a limited number of (typically <1000) peptides and aim to identify T cells responsive to them. For instance, Wang et al. characterized the immunopeptidome of HLA-DR15 and identified self-epitopes and their microbial derived mimics as autoantigens in Multiple Sclerosis28. On the other hand, TCR-directed approaches start with key TCRs and screen them against large epitope libraries (up to 10^7). We have recently identified novel T1D autoantigens using cell-based epitope libraries that were derived from a mouse pancreatic islet immunopeptidomics study62,101. In both cases, the knowledge of the peptides that were actually presented on MHC/HLA augmented antigen discovery by narrowing down the possible universe of epitopes recognized by T cells under investigation28,102,103. In case of autoimmunity, this is especially important, as the scale of potential self-epitopes is genome-wide. In addition to the ~20,000 annotated coding genes, there are >10,000 unannotated or non-canonical open reading frames that contribute to the immunopeptidome. Furthermore, epitopes with PTMs add to this landscape of potential autoantigens. Immunopeptidomics can be used to scale down the number of epitopes under investigation, allowing better throughput and more targeted antigen discovery. In the next section, we will describe how immunopeptidomics approaches have helped autoantigen discovery in specific cases of autoimmune diseases: T1D, SLE, and RA.
Type 1 Diabetes
T1D or Autoimmune Diabetes, is a chronic disease caused by insulin deficiency due to the destruction of the insulin-producing β cells in the pancreatic islets of Langerhans104,105. Autoantibodies against insulin, the 65-kDa form of glutamic acid decarboxylase (GAD65), insulinoma-associated protein 2 (IA-2), and zinc transporter 8 (ZnT8) are associated with T1D but their role in the pathophysiology of the disease is not clear106. It has been shown that autoreactive CD4+ and CD8+ T cells infiltrate the pancreas and mediate destruction of β cells. CD4+ T cells can propagate a pro-inflammatory environment through cytokine secretion and enhancing the function of cytotoxic CD8 + T cells, which can directly kill β cells. T1D can be modeled in NOD mice, which share several key features of disease including the presence of islet autoantibodies and infiltration of autoreactive CD4+ and CD8 + T cells in islets107,108,109,110. Islet-infiltrating T cells in NOD mice and T1D are known to recognize β cell autoantigens, many of which are overlapping. The restricted MHC diversity as well as availability of samples and reproducibility of disease course have allowed robust immunopeptidomics studies in NOD mice57,111. In contrast, the high HLA heterogeneity and limited access to viable β cells have impeded similar studies in humans. Islets harvested from cadaveric donors with T1D have very few β cells remaining, and those from donors without T1D have naturally low levels of HLA expression in the absence of inflammation, making direct detection of presented peptides challenging. Moreover, even in inflamed islet, class II HLA expression on β cells is low112,113,114,115. Therefore, the characterization of the HLA bound peptides from human β cells has been limited106,116. To circumvent these issues, approaches such as stably transfected human non β cell lines expressing specific autoantigen(s) and cell surface HLA allotypes of interest117,118, or human β cell lines generated by targeted oncogenesis116 have been used. A recent finding in T1D was the presence of hybrid insulin peptides (HIPs), which consist of epitopes generated by post-translational splicing of Insulin with other proteins. Studies have shown that HIPs are autoantigens for pathogenic CD4+ T cells in the human T1D and in NOD mice119,120, confirming the notion that PTM epitopes are key autoantigens121,122. Importantly, while the initial experiments with HIPs used synthetic peptides, their presence in the proteomes and immunopeptidomes derived from β cells has reaffirmed that HIP formation and recognition is a natural process that occurs in T1D119. Subsequent studies based off of these results have led to the identification of novel HIPs as diabetogenic epitopes.
We wish to highlight two recent studies that have effectively combined immunopeptidomics and antigen discovery approaches to identify novel autoantigens in T1D. Gonzalez-Duque et al. performed class I HLA immunopeptidomics on a human β cell line, ECN90, and on islets, and identified ~3000 peptides including native peptides, PTM epitopes, splice variants, and transpeptidation products (which are similar to HIPs, but their existence is still debated). Using synthetic peptides and peptide-MHC multimers, the authors identified several novel autoantigens including insulin gene enhancer protein ISL-1 and UCN3. T cells recognizing these autoantigens were shown to be enriched in the pancreata of T1D donors as compared with non-diabetic donors116. In the second study, Wan et al. performed class II MHC immunopeptidomics on pancreatic islets and draining lymph nodes in NOD mice, and identified >4000 peptides bound to I-Ag762. They also identified many PTM epitopes, splice variants, and HIPs. Using this immunopeptidomics dataset, our group built epitope libraries presenting >4000 epitopes in I-Ag7, and identified targets of islet-infiltrating T cells de novo, and found a predominance of HIP-reactive T cells101. These studies exemplify that combining immunopeptidomics with antigen discovery can be a powerful strategy for identifying autoantigens.
Systemic Lupus Erythematosus
SLE is a multisystem, chronic autoimmune disease involving a complex interaction of impaired apoptotic clearance, complement activation, and immune complex formation which leads to dysregulated innate and adaptive immunity123,124,125,126. SLE is characterized by the presence of autoantibodies to nuclear and cytoplasmic antigens127. While the importance of B cells and anti-nuclear antibodies in SLE pathogenesis is appreciated, tissue infiltrating T cells also play a key role128. The antigenic landscape of autoreactive T cells in SLE is poorly defined, with a handful of known autoantigens, such as histones, described to date129,130. Interestingly, histones and other nuclear proteins are broadly modified post-translationally131, but whether these PTMs lead to immunogenic epitopes is not known. Proteomics profiling of tissues and plasma in SLE patients and mouse models have shown changes in the soluble proteome associated with inflammation and immune dysregulation132,133. Antibodies to canonical autoantigens in SLE such as like Smith, RO, La, and histones, have been detected in patient sera, and serve as disease biomarkers134,135. First identification of T cell specificities in SLE came from curating a small list of potential autoantigens from proteomics datasets. Critically, the link between SLE proteomes and SLE immunopeptidomes is missing, largely due to the lack of immunopeptidomics data from mice or humans.
SLE, unlike T1D, has a high level of heterogeneity in disease course, target tissues, and environmental triggers, therefore making it challenging to hone in on the key antigen-presenting populations. Only a small number of studies have reported immunopeptidomes in mouse models of SLE. In the early 2000s, Freed et al. characterized the peptides eluted from class II MHC from spleens in a SLE-prone mouse model, I-Ak or I-Ek alleles in MRL/lpr mice. A very small number ( < 20) peptides were detected, including some from potential SLE autoantigens such as histones136. The study only uncovered a small number of peptides due to technical limitations like lower limit of detection and low abundance peptides137. We have recently performed immunopeptidomics on the kidneys of MRL/lpr mice, which is the primary pathologic site in SLE. We identified >3000 epitopes presented on I-Ek in kidneys of MRL/lpr mice, and has used interaction language models to predict potential immunogens. In concordance with the previous reports, we did indeed detect peptides derived from histones and ribosomal proteins138. In addition, we have developed an algorithm that will be able to predict HLA restriction of peptides previously not studied, this will advance our ability to tackle the HLA diversity. We believe that with the recent technological advances in immunopeptidomics, time is ripe to deploy it for autoantigen discovery in SLE. However, several key considerations still need to be taken into account, including the HLA diversity in humans, availability of kidney tissue from patients, and possible PTMs. Moreover, profiling the immunopeptidome will need to be followed by experimental validation of their immunogenicity in mice and humans. The application of immunopeptidomics will be essential for identifying autoantigens in SLE that will serve as diagnostic and therapeutic targets.
Rheumatoid Arthritis
RA is a systemic, inflammatory autoimmune disease, characterized by immune infiltration into the synovial joints, leading to varying degrees of functional impairment among patients139. A prognostic hallmark of RA is the presence of autoantibodies that recognize self-proteins harboring PTMs like citrullination, homocitrullination (carbamylation), and acetylation140,141,142. Genetic association with certain HLA-DR alleles and the presence of anti-citrullinated protein antibodies (ACPAs), suggests a pathophysiological role of CD4+ T cells in disease143,144. While CD4+ T cell infiltration in the synovial tissue is characteristic of RA, the precise autoantigens recognized by them are poorly defined145,146. Most studies in RA have focused predominantly on the HLA-DR immunopeptidome, given that multiple HLA-DR molecules are strongly associated with the disease. Among the most well-described risk alleles for RA is the shared epitope (SE), a set of HLA-DRB1 alleles containing a consensus amino acid sequence in residues 70-74 of the HLA-DRβ chain147. SE positivity strongly correlates with ACPA positivity and is associated with earlier onset of RA, increased disease severity, and higher mortality147,148. SE-containing HLA-DR alleles are thought to have enhanced presentation of arthritogenic antigens, including citrullinated peptides, leading to selection autoreactive T cell repertoires149,150,151 and promotion of ACPA formation76.
In a 2010 study, 1427 HLA-DR-presented peptides, derived from 166 source proteins were identified in the synovia of two RA patients152. Another study examining clinical samples of synovial tissue, synovial fluid mononuclear cells, and peripheral blood mononuclear cells identified 1593 peptides originating from 870 source proteins153. A key mechanistic insight into how certain HLA alleles influence the genetic risk was obtained through immunopeptidomics studies comparing 962 unique peptides bound to strongly RA-associated DRB1*01:01, DRB1*04:01, and DRB1*10:01 alleles and non-RA-associated DRB1*15:01 allele154. It was found that the peptide repertoires differed largely in terms of size, protein origin, composition, and affinity, with only about 10% overlap among RA-associated allotypes. Such empirical data on allelic binding preferences will enhance bioinformatics-based prediction tools that infer peptide repertoires. For example, Darrah et al. used the NetMHCII-2.3 binding prediction algorithm combined with proteolytic mapping to predict binding affinities for peptides derived from native and citrullinated antigens to RA-associated SE alleles (i.e., DRB*01:01, *04:01, *04:05, and *10:01). They demonstrated that structural changes induced by citrullination alter susceptibility to proteolytic cleavage, thus modulating antigen processing and revealing cryptic epitopes155. Additionally, Kaabinejadian et al. used MHCMotifDecon on existing immunopeptidomic datasets and found that the secondary DR alleles (HLA-DRB3, DRB4, and DRB5), often overlooked due to their strong linkage disequilibrium with the primary HLA-DRB1 allele, contributed significantly to the HLA-DR repertoire. They posit that secondary DR alleles, which display non-redundant and complementary peptide repertoires, warrant regard as functionally independent alleles in future studies156. This mechanistic understanding of RA susceptibility and HLA-associated variability would not have been possible without immunopeptidomics.
Translational potential of immunopeptidomics
Overall, immunopeptidomics has provided valuable insights into the pathogenesis of autoimmune diseases and has helped to define key autoantigens. While immunopeptidomics studies in autoimmune diseases are often not directly translatable, they have tremendous potential to fuel diagnostic and therapeutic approaches. Generation of large datasets of MHC/HLA-bound peptides have led to the advances in computational tools to predict epitope binding138,157,158. These algorithms can be then used to predict potential autoantigens. Immunopeptidomics datasets have directly fueled systematic antigen discovery approaches which are leading to using antigen-specific TCR repertoires as diagnostic tools. Moreover, novel autoantigens can be directly used as immunogens to induce tolerance or as targets for immunomodulatory strategies. Furthermore, immunopeptidomics has revealed mechanistic aspects of development of autoimmunity, such as cross-reactivity with microbial epitopes, changes in immune landscapes associated with disease states, and PTM-driven alterations in HLA binding and T cell recognition.
Summary and future directions
Identifying the peptides that are presented on MHC/HLA has led to several key advances in autoimmunity including autoantigen identification, confirmation of the biological relevance for HIPs, and to an increasing appreciation for PTM epitopes as pathogenic. Several key considerations remain in the design and utility of immunopeptidomics studies, as highlighted in Fig. 1. As the experimental techniques to elute and detect peptides and computational tools to analyze datasets advance, we envision that the numbers of identified peptides will continue growing exponentially. This will enhance our understanding of the pathogenic antigens across autoimmune disorders. In summary, as immunopeptidomics opens this chamber of secrets, a treasure trove of autoantigens will be discovered.
References
Davis, M. M. & Bjorkman, P. J. T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402 (1988).
Hesslein, D. G. & Schatz, D. G. Factors and forces controlling V(D)J recombination. Adv. Immunol. 78, 169–232 (2001).
Kisielow, P. & von Boehmer, H. Development and selection of T cells: facts and puzzles. Adv. Immunol. 58, 87–209 (1995).
Lanier, L. L. et al. The gamma T-cell antigen receptor. J. Clin. Immunol. 7, 429–440 (1987).
Adams, E. J., Gu, S. & Luoma, A. M. Human gamma delta T cells: Evolution and ligand recognition. Cell Immunol. 296, 31–40 (2015).
Chicz, R. M. et al. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature 358, 764–768 (1992).
Engelhard, V. H. Structure of peptides associated with class I and class II MHC molecules. Annu Rev. Immunol. 12, 181–207 (1994).
Roche, P. A. & Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203–216 (2015).
Truong, H. V. & Sgourakis, N. G. Dynamics of MHC-I molecules in the antigen processing and presentation pathway. Curr. Opin. Immunol. 70, 122–128 (2021).
Schmidt, E. M. et al. Ctla-4 controls regulatory T cell peripheral homeostasis and is required for suppression of pancreatic islet autoimmunity. J. Immunol. 182, 274–282 (2009).
Wang, J. et al. PD-1 deficiency results in the development of fatal myocarditis in MRL mice. Int Immunol. 22, 443–452 (2010).
Blum, J. S., Wearsch, P. A. & Cresswell, P. Pathways of antigen processing. Annu Rev. Immunol. 31, 443–473 (2013).
Pishesha, N., Harmand, T. J. & Ploegh, H. L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 22, 751–764 (2022).
Rock, K. L., Reits, E. & Neefjes, J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol. 37, 724–737 (2016).
Cosgrove, D. et al. Mice lacking MHC class II molecules. Cell 66, 1051–1066 (1991).
Grusby, M. J. et al. Mice lacking major histocompatibility complex class I and class II molecules. Proc. Natl Acad. Sci. USA 90, 3913–3917 (1993).
Zijlstra, M. et al. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature 344, 742–746 (1990).
Spurgin, L. G. & Richardson, D. S. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. Biol. Sci. 277, 979–988 (2010).
Robinson, J. H. & Delvig, A. A. Diversity in MHC class II antigen presentation. Immunology 105, 252–262 (2002).
Slieker, R. C. et al. Reassessing human MHC-I genetic diversity in T cell studies. Sci. Rep. 14, 7966 (2024).
Lenz, T. L. et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat. Genet 47, 1085–1090 (2015).
Ishigaki, K. et al. HLA autoimmune risk alleles restrict the hypervariable region of T cell receptors. Nat. Genet 54, 393–402 (2022).
Noble, J. A. & Valdes, A. M. Genetics of the HLA region in the prediction of type 1 diabetes. Curr. Diab Rep. 11, 533–542 (2011).
Takaki, T. et al. HLA-A*0201-restricted T cells from humanized NOD mice recognize autoantigens of potential clinical relevance to type 1 diabetes. J. Immunol. 176, 3257–3265 (2006).
Noble, J. A. & Erlich, H. A. Genetics of type 1 diabetes. Cold Spring Harb. Perspect. Med 2, a007732 (2012).
Eyre, S. et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet 44, 1336–1340 (2012).
Inoue, M. et al. Carriers of HLA-DRB1*04:05 have a better clinical response to abatacept in rheumatoid arthritis. Sci. Rep. 13, 15250 (2023).
Wang, J. et al. HLA-DR15 Molecules Jointly Shape an Autoreactive T Cell Repertoire in Multiple Sclerosis. Cell 183, 1264–1281.e1220 (2020).
Simmons, K. M. et al. Failed Genetic Protection: Type 1 Diabetes in the Presence of HLA-DQB1*06:02. Diabetes 69, 1763–1769 (2020).
Jersild, C. et al. Histocompatibility determinants in multiple sclerosis, with special reference to clinical course. Lancet 2, 1221–1225 (1973).
Martin, R., Sospedra, M., Eiermann, T. & Olsson, T. Multiple sclerosis: doubling down on MHC. Trends Genet 37, 784–797 (2021).
Moutsianas, L. et al. Class II HLA interactions modulate genetic risk for multiple sclerosis. Nat. Genet 47, 1107–1113 (2015).
Wieczorek, M. et al. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol. 8, 292 (2017).
Radwan, J., Babik, W., Kaufman, J., Lenz, T. L. & Winternitz, J. Advances in the Evolutionary Understanding of MHC Polymorphism. Trends Genet 36, 298–311 (2020).
Sommer, S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool. 2, 16 (2005).
Pereyra, F. et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330, 1551–1557 (2010).
Kaslow, R. A. et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med 2, 405–411 (1996).
Chen, H. et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat. Immunol. 13, 691–700 (2012).
Goulder, P. J. et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412, 334–338 (2001).
McMichael, A. & Bowness, P. HLA-B27: natural function and pathogenic role in spondyloarthritis. Arthritis Res 4, S153–158 (2002).
Suri, A., Walters, J. J., Gross, M. L. & Unanue, E. R. Natural peptides selected by diabetogenic DQ8 and murine I-A(g7) molecules show common sequence specificity. J. Clin. Invest 115, 2268–2276 (2005).
Todd, J. A., Bell, J. I. & McDevitt, H. O. H. L. A.-D. Q. beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 329, 599–604 (1987).
Latek, R. R. et al. Structural basis of peptide binding and presentation by the type I diabetes-associated MHC class II molecule of NOD mice. Immunity 12, 699–710 (2000).
Gioia, L. et al. Position β57 of I-A(g7) controls early anti-insulin responses in NOD mice, linking an MHC susceptibility allele to type 1 diabetes onset. Sci Immunol 4, https://doi.org/10.1126/sciimmunol.aaw6329 (2019).
Glaeser, R. M., Downing, K. H. & Jap, B. K. What spectroscopy can still tell us about the secondary structure of bacteriorhodopsin. Biophys. J. 59, 934–938 (1991).
Dausset, J. Iso-leuko-antibodies. Acta Haematol. 20, 156–166 (1958).
Benacerraf, B. & McDevitt, H. O. Histocompatibility-linked immune response genes. Science 175, 273–279 (1972).
Dausset, J., Rapaport, F. T., Colombani, J. & Feingold, N. A leucocyte group and its relationship to tissue histocompatibility in man. Transplantation 3, 701–705 (1965).
Green, I., Paul, W. E. & Benacerraf, B. Genetic control of immunological responsiveness in guinea pigs to 2,4-dinitrophenyl conjugates of poly-L-arginine, protamine, and poly-L-ornithine. Proc. Natl Acad. Sci. USA 64, 1095–1102 (1969).
McDevitt, H. O. & Chinitz, A. Genetic control of the antibody response: relationship between immune response and histocompatibility (H-2) type. Science 163, 1207–1208 (1969).
McDevitt, H. O. & Sela, M. Genetic control of the antibody response. I. Demonstration of determinant-specific differences in response to synthetic polypeptide antigens in two strains of inbred mice. J. Exp. Med. 122, 517–531 (1965).
Tyan, M. L., McDevitt, H. O. & Herzenberg, L. A. Genetic control of the antibody response to a synthetic polypeptide: transfer of response with spleen cells or lymphoid precursors. Transpl. Proc. 1, 548–550 (1969).
Zinkernagel, R. M. & Doherty, P. C. Cytotoxic thymus-derived lymphocytes in cerebrospinal fluid of mice with lymphocytic choriomeningitis. J. Exp. Med 138, 1266–1269 (1973).
Bjorkman, P. J. et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329, 506–512 (1987).
Garcia, K. C. et al. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 274, 209–219 (1996).
Prensner, J. R. et al. What Can Ribo-Seq, Immunopeptidomics, and Proteomics Tell Us About the Noncanonical Proteome? Mol. Cell Proteom. 22, 100631 (2023).
Lichti, C. F. & Wan, X. Using mass spectrometry to identify neoantigens in autoimmune diseases: The type 1 diabetes example. Semin Immunol. 66, 101730 (2023).
Stopfer, L. E., D’Souza, A. D. & White, F. M. 1,2,3, MHC: a review of mass-spectrometry-based immunopeptidomics methods for relative and absolute quantification of pMHCs. Immunooncol Technol. 11, 100042 (2021).
Leddy, O. K., White, F. M. & Bryson, B. D. Leveraging Immunopeptidomics To Study and Combat Infectious Disease. mSystems 6, e0031021 (2021).
Hunt, D. F. et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 (1992).
Li, X. et al. A microfluidics-enabled automated workflow of sample preparation for MS-based immunopeptidomics. Cell Rep. Methods 3, 100479 (2023).
Wan, X. et al. The MHC-II peptidome of pancreatic islets identifies key features of autoimmune peptides. Nat. Immunol. 21, 455–463 (2020).
Klaeger, S. et al. Optimized Liquid and Gas Phase Fractionation Increases HLA-Peptidome Coverage for Primary Cell and Tissue Samples. Mol. Cell Proteom. 20, 100133 (2021).
Hoenisch Gravel, N. et al. TOF(IMS) mass spectrometry-based immunopeptidomics refines tumor antigen identification. Nat. Commun. 14, 7472 (2023).
Munday, P. R. et al. Immunolyser: A web-based computational pipeline for analysing and mining immunopeptidomic data. Comput Struct. Biotechnol. J. 21, 1678–1687 (2023).
Huang, X. et al. The SysteMHC Atlas v2.0, an updated resource for mass spectrometry-based immunopeptidomics. Nucleic Acids Res 52, D1062–D1071 (2024).
Ouspenskaia, T. et al. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat. Biotechnol. 40, 209–217 (2022).
Holly, J. & Yewdell, J. W. Game of Omes: ribosome profiling expands the MHC-I immunopeptidome. Curr. Opin. Immunol. 83, 102342 (2023).
Adams, C. et al. Fragment ion intensity prediction improves the identification rate of non-tryptic peptides in timsTOF. Nat. Commun. 15, 3956 (2024).
Gessulat, S. et al. Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods 16, 509–518 (2019).
Declercq, A. et al. MS(2)Rescore: Data-Driven Rescoring Dramatically Boosts Immunopeptide Identification Rates. Mol. Cell Proteom. 21, 100266 (2022).
Yang, K. L. et al. MSBooster: improving peptide identification rates using deep learning-based features. Nat. Commun. 14, 4539 (2023).
Schuster, H. et al. A tissue-based draft map of the murine MHC class I immunopeptidome. Sci. Data 5, 180157 (2018).
Ramazi, S. & Zahiri, J. Posttranslational modifications in proteins: resources, tools and prediction methods. Database (Oxford) 2021, https://doi.org/10.1093/database/baab012 (2021).
Jennings, E. Q., Fritz, K. S. & Galligan, J. J. Biochemical genesis of enzymatic and non-enzymatic post-translational modifications. Mol. Asp. Med 86, 101053 (2022).
Haro, I., Sanmartí, R. & Gómara, M. J. Implications of Post-Translational Modifications in Autoimmunity with Emphasis on Citrullination, Homocitrullination and Acetylation for the Pathogenesis, Diagnosis and Prognosis of Rheumatoid Arthritis. Int J Mol Sci 23, https://doi.org/10.3390/ijms232415803 (2022).
Chen, Z. et al. Systematic Characterization of Lysine Post-translational Modification Sites Using MUscADEL. Methods Mol. Biol. 2499, 205–219 (2022).
Li, Z. et al. dbPTM in 2022: an updated database for exploring regulatory networks and functional associations of protein post-translational modifications. Nucleic Acids Res 50, D471–D479 (2022).
Yan, Y. et al. MIND-S is a deep-learning prediction model for elucidating protein post-translational modifications in human diseases. Cell Rep. Methods 3, 100430 (2023).
Skipper, J. C. et al. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med 183, 527–534 (1996).
Engelhard, V. H., Altrich-Vanlith, M., Ostankovitch, M. & Zarling, A. L. Post-translational modifications of naturally processed MHC-binding epitopes. Curr. Opin. Immunol. 18, 92–97 (2006).
Zarling, A. L. et al. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc. Natl Acad. Sci. USA 103, 14889–14894 (2006).
Liepe, J. et al. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 (2016).
Faridi, P. et al. A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci Immunol 3, https://doi.org/10.1126/sciimmunol.aar3947 (2018).
Mylonas, R. et al. Estimating the Contribution of Proteasomal Spliced Peptides to the HLA-I Ligandome. Mol. Cell Proteom. 17, 2347–2357 (2018).
Mei, S. et al. Immunopeptidomic analysis reveals that deamidated HLA-bound peptides arise predominantly from deglycosylated precursors. Mol. Cell Proteom. 19, 1236–1247 (2020).
Dalet, A. et al. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc. Natl Acad. Sci. USA 108, E323–E331 (2011).
Rolfs, Z., Solntsev, S. K., Shortreed, M. R., Frey, B. L. & Smith, L. M. Global Identification of Post-Translationally Spliced Peptides with Neo-Fusion. J. Proteome Res 18, 349–358 (2019).
Sandalova, T., Sala, B. M. & Achour, A. Structural aspects of chemical modifications in the MHC-restricted immunopeptidome; Implications for immune recognition. Front Chem. 10, 861609 (2022).
Prensner, J. R. et al. What can Ribo-seq and proteomics tell us about the non-canonical proteome? bioRxiv, https://doi.org/10.1101/2023.05.16.541049 (2023).
Bedran, G. et al. The Immunopeptidome from a Genomic Perspective: Establishing the Noncanonical Landscape of MHC Class I-Associated Peptides. Cancer Immunol. Res 11, 747–762 (2023).
Mangalaparthi, K. K. et al. Digging deeper into the immunopeptidome: characterization of post-translationally modified peptides presented by MHC I. J. Proteins Proteom. 12, 151–160 (2021).
Santambrogio, L. Autoimmunity to the modified self. Science 379, 1092–1093 (2023).
Houde, D., Peng, Y., Berkowitz, S. A. & Engen, J. R. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol. Cell Proteom. 9, 1716–1728 (2010).
Zhai, Y. & Zhu, P. Post-translationally modified neoantigens: Promising targets for diagnostic strategy of autoimmune diseases. Clin. Transl. Med 13, e1373 (2023).
Cole, D. K. et al. Modification of MHC anchor residues generates heteroclitic peptides that alter TCR binding and T cell recognition. J. Immunol. 185, 2600–2610 (2010).
Raposo, B. et al. T cells specific for post-translational modifications escape intrathymic tolerance induction. Nat. Commun. 9, 353 (2018).
Petersen, J., Purcell, A. W. & Rossjohn, J. Post-translationally modified T cell epitopes: immune recognition and immunotherapy. J. Mol. Med. (Berl.) 87, 1045–1051 (2009).
Yi, X. et al. caAtlas: An immunopeptidome atlas of human cancer. iScience 24, 103107 (2021).
Joglekar, A. V. & Li, G. T cell antigen discovery. Nat. Methods 18, 873–880 (2021).
Zdinak, P. M. et al. De novo identification of CD4(+) T cell epitopes. Nat. Methods 21, 846–856 (2024).
Stadinski, B. D. et al. Chromogranin A is an autoantigen in type 1 diabetes. Nat. Immunol. 11, 225–231 (2010).
Wiles, T. A. et al. An insulin-IAPP hybrid peptide is an endogenous antigen for CD4 T cells in the non-obese diabetic mouse. J. Autoimmun. 78, 11–18 (2017).
Bluestone, J. A., Herold, K. & Eisenbarth, G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature 464, 1293–1300 (2010).
Kahanovitz, L., Sluss, P. M. & Russell, S. J. Type 1 Diabetes - A Clinical Perspective. Point Care 16, 37–40 (2017).
Purcell, A. W., Sechi, S. & DiLorenzo, T. P. The evolving landscape of autoantigen discovery and characterization in type 1 diabetes. Diabetes 68, 879–886 (2019).
Anderson, M. S. & Bluestone, J. A. The NOD mouse: a model of immune dysregulation. Annu Rev. Immunol. 23, 447–485 (2005).
Pearson, J. A., Wong, F. S. & Wen, L. The importance of the Non Obese Diabetic (NOD) mouse model in autoimmune diabetes. J. Autoimmun. 66, 76–88 (2016).
Wong, F. S., Visintin, I., Wen, L., Flavell, R. A. & Janeway, C. A. Jr. CD8 T cell clones from young nonobese diabetic (NOD) islets can transfer rapid onset of diabetes in NOD mice in the absence of CD4 cells. J. Exp. Med 183, 67–76 (1996).
Unanue, E. R. Antigen presentation in the autoimmune diabetes of the NOD mouse. Annu Rev. Immunol. 32, 579–608 (2014).
Dudek, N. L. et al. Constitutive and inflammatory immunopeptidome of pancreatic β-cells. Diabetes 61, 3018–3025 (2012).
Quesada-Masachs, E. et al. Upregulation of HLA class II in pancreatic beta cells from organ donors with type 1 diabetes. Diabetologia 65, 387–401 (2022).
Russell, M. A. et al. HLA Class II Antigen Processing and Presentation Pathway Components Demonstrated by Transcriptome and Protein Analyses of Islet β-Cells From Donors With Type 1 Diabetes. Diabetes 68, 988–1001 (2019).
Zhao, Y. et al. Mouse pancreatic beta cells express MHC class II and stimulate CD4(+) T cells to proliferate. Eur. J. Immunol. 45, 2494–2503 (2015).
Harris, P. E. et al. Effect of interferon alpha on MHC class II gene expression in ex vivo human islet tissue. Biochim Biophys. Acta 1762, 627–635 (2006).
Gonzalez-Duque, S. et al. Conventional and Neo-antigenic Peptides Presented by β Cells Are Targeted by Circulating Naïve CD8+ T Cells in Type 1 Diabetic and Healthy Donors. Cell Metab. 28, 946–960.e946 (2018).
Kronenberg, D. et al. Circulating preproinsulin signal peptide-specific CD8 T cells restricted by the susceptibility molecule HLA-A24 are expanded at onset of type 1 diabetes and kill β-cells. Diabetes 61, 1752–1759 (2012).
Skowera, A. et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J. Clin. Invest 118, 3390–3402 (2008).
Wiles, T. A. et al. Identification of Hybrid Insulin Peptides (HIPs) in Mouse and Human Islets by Mass Spectrometry. J. Proteome Res 18, 814–825 (2019).
Delong, T. et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 351, 711–714 (2016).
Doyle, H. A. & Mamula, M. J. Posttranslational modifications of self-antigens. Ann. N. Y Acad. Sci. 1050, 1–9 (2005).
Dunne, J. L., Overbergh, L., Purcell, A. W. & Mathieu, C. Posttranslational modifications of proteins in type 1 diabetes: the next step in finding the cure? Diabetes 61, 1907–1914 (2012).
Sullivan, K. E. Genetics of systemic lupus erythematosus. Clin. Implic. Rheum. Dis. Clin. North Am. 26, 229–256 (2000). v-vi.
Moser, K. L., Kelly, J. A., Lessard, C. J. & Harley, J. B. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 10, 373–379 (2009).
Tsokos, G. C. & Kammer, G. M. Molecular aberrations in human systemic lupus erythematosus. Mol. Med Today 6, 418–424 (2000).
Faurschou, M., Starklint, H., Halberg, P. & Jacobsen, S. Prognostic factors in lupus nephritis: diagnostic and therapeutic delay increases the risk of terminal renal failure. J. Rheumatol. 33, 1563–1569 (2006).
Karrar, S. & Cunninghame Graham, D. S. Abnormal B cell development in systemic lupus erythematosus: what the genetics tell us. Arthritis Rheumatol. 70, 496–507 (2018).
Tsokos, G. C., Lo, M. S., Costa Reis, P. & Sullivan, K. E. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 12, 716–730 (2016).
Riemekasten, G. et al. Identification and characterization of SmD183-119-reactive T cells that provide T cell help for pathogenic anti-double-stranded DNA antibodies. Arthritis Rheum. 48, 475–485 (2003).
Riemekasten, G. & Hahn, B. H. Key autoantigens in SLE. Rheumatol. (Oxf.) 44, 975–982 (2005).
Navarro Quiroz, E. et al. Epigenetic Mechanisms and Posttranslational Modifications in Systemic Lupus Erythematosus. Int J Mol Sci 20, https://doi.org/10.3390/ijms20225679 (2019).
Madda, R., Lin, S. C., Sun, W. H. & Huang, S. L. Plasma proteomic analysis of systemic lupus erythematosus patients using liquid chromatography/tandem mass spectrometry with label-free quantification. PeerJ 6, e4730 (2018).
Reynolds, J. A., Li, Y., Herlitz, L., Mohan, C. & Putterman, C. Novel biomarker discovery through comprehensive proteomic analysis of lupus mouse serum. J. Autoimmun. 142, 103134 (2024).
Nicolaou, O. et al. Biomarkers of systemic lupus erythematosus identified using mass spectrometry-based proteomics: a systematic review. J. Cell Mol. Med 21, 993–1012 (2017).
Tesch, S. et al. Identification and characterization of antigen-specific CD4(+) T cells targeting renally expressed antigens in human lupus nephritis with two independent methods. Sci. Rep. 10, 21312 (2020).
Freed, J. H., Marrs, A., VanderWall, J., Cohen, P. L. & Eisenberg, R. A. MHC class II-bound self peptides from autoimmune MRL/lpr mice reveal potential T cell epitopes for autoantibody production in murine systemic lupus erythematosus. J. Immunol. 164, 4697–4705 (2000).
Hunt, D. F. et al. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science 256, 1817–1820 (1992).
Omelchenko, A. A. et al. Sliding Window INteraction Grammar (SWING): a generalized interaction language model for peptide and protein interactions. bioRxiv, https://doi.org/10.1101/2024.05.01.592062 (2024).
Bullock, J. et al. Rheumatoid arthritis: a brief overview of the treatment. Med Princ. Pr. 27, 501–507 (2018).
van Delft, M. A. M. & Huizinga, T. W. J. An overview of autoantibodies in rheumatoid arthritis. J. Autoimmun. 110, 102392 (2020).
de Brito Rocha, S., Baldo, D. C. & Andrade, L. E. C. Clinical and pathophysiologic relevance of autoantibodies in rheumatoid arthritis. Adv. Rheumatol. 59, 2 (2019).
Gavrilă, B. I., Ciofu, C. & Stoica, V. Biomarkers in Rheumatoid Arthritis, what is new? J. Med Life 9, 144–148 (2016).
Cope, A. P., Schulze-Koops, H. & Aringer, M. The central role of T cells in rheumatoid arthritis. Clin. Exp. Rheumatol. 25, S4–S11 (2007).
van Drongelen, V. & Holoshitz, J. Human leukocyte antigen-disease associations in rheumatoid arthritis. Rheum. Dis. Clin. North Am. 43, 363–376 (2017).
Chemin, K., Gerstner, C. & Malmström, V. Effector functions of CD4+ T cells at the site of local autoimmune inflammation-lessons from rheumatoid arthritis. Front Immunol. 10, 353 (2019).
Cope, A. P. T cells in rheumatoid arthritis. Arthritis Res Ther. 10, S1 (2008).
Holoshitz, J. The rheumatoid arthritis HLA-DRB1 shared epitope. Curr. Opin. Rheumatol. 22, 293–298 (2010).
Romão, V. C. & Fonseca, J. E. Etiology and Risk Factors for Rheumatoid Arthritis: A State-of-the-Art Review. Front Med (Lausanne) 8, 689698 (2021).
Wucherpfennig, K. W. & Strominger, J. L. Selective binding of self peptides to disease-associated major histocompatibility complex (MHC) molecules: a mechanism for MHC-linked susceptibility to human autoimmune diseases. J. Exp. Med. 181, 1597–1601 (1995).
Bhayani, H. R. & Hedrick, S. M. The role of polymorphic amino acids of the MHC molecule in the selection of the T cell repertoire. J. Immunol. 146, 1093–1098 (1991).
Nguyen, H. & James, E. A. Immune recognition of citrullinated epitopes. Immunology 149, 131–138 (2016).
Seward, R. J., Drouin, E. E., Steere, A. C. & Costello, C. E. Peptides presented by HLA-DR molecules in synovia of patients with rheumatoid arthritis or antibiotic-refractory Lyme arthritis. Mol. Cell Proteom. 10, M110.002477 (2011).
Wang, Q. et al. Immunogenic HLA-DR-presented self-peptides identified directly from clinical samples of synovial tissue, synovial fluid, or peripheral blood in patients with rheumatoid arthritis or lyme arthritis. J. Proteome Res 16, 122–136 (2017).
Scholz, E. et al. A Comparative Analysis of the Peptide Repertoires of HLA-DR Molecules Differentially Associated With Rheumatoid Arthritis. Arthritis Rheumatol. 68, 2412–2421 (2016).
Curran, A. M. et al. Citrullination modulates antigen processing and presentation by revealing cryptic epitopes in rheumatoid arthritis. Nat. Commun. 14, 1061 (2023).
Kaabinejadian, S. et al. Accurate MHC motif deconvolution of immunopeptidomics data reveals a significant contribution of DRB3, 4 and 5 to the Total DR Immunopeptidome. Front Immunol. 13, 835454 (2022).
Nielsen, M. et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2, e796 (2007).
Nielsen, M. et al. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol. 4, e1000107 (2008).
Maggi, J. et al. Isolation of HLA-DR-naturally presented peptides identifies T-cell epitopes for rheumatoid arthritis. Ann. Rheum. Dis. 81, 1096–1105 (2022).
Xu, Z. et al. Integrative Proteomics and N-glycoproteomics analyses of rheumatoid arthritis synovium reveal immune-associated glycopeptides. Mol. Cell Proteom. 22, 100540 (2023).
Azoury, M. E. et al. Peptides Derived From Insulin Granule Proteins Are Targeted by CD8(+) T Cells Across MHC Class I Restrictions in Humans and NOD Mice. Diabetes 69, 2678–2690 (2020).
Dolton, G. et al. HLA A*24:02-restricted T cell receptors cross-recognize bacterial and preproinsulin peptides in type 1 diabetes. J Clin Invest 134, https://doi.org/10.1172/jci164535 (2024).
Anderson, A. M. et al. Human islet T cells are highly reactive to preproinsulin in type 1 diabetes. Proc Natl Acad Sci USA 118, https://doi.org/10.1073/pnas.2107208118 (2021).
Unger, W. W. et al. Discovery of low-affinity preproinsulin epitopes and detection of autoreactive CD8 T-cells using combinatorial MHC multimers. J. Autoimmun. 37, 151–159 (2011).
Toma, A. et al. Recognition of a subregion of human proinsulin by class I-restricted T cells in type 1 diabetic patients. Proc. Natl Acad. Sci. USA 102, 10581–10586 (2005).
Landry, L. G. et al. Proinsulin-Reactive CD4 T Cells in the Islets of Type 1 Diabetes Organ Donors. Front Endocrinol. (Lausanne) 12, 622647 (2021).
van Lummel, M. et al. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes 63, 237–247 (2014).
Tran, M. T. et al. A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8. J. Biol. Chem. 300, 107612 (2024).
Callebaut, A. et al. CD4+ T Cells From Individuals With Type 1 Diabetes Respond to a Novel Class of Deamidated Peptides Formed in Pancreatic Islets. Diabetes 73, 728–742 (2024).
Arribas-Layton, D. et al. Hybrid Insulin Peptides Are Recognized by Human T Cells in the Context of DRB1*04:01. Diabetes 69, 1492–1502 (2020).
Berner, B. R. et al. Phenotypic and functional analysis of EBV-specific memory CD8 cells in SLE. Cell Immunol. 235, 29–38 (2005).
Ito, H. et al. Analysis of T cell responses to the beta 2-glycoprotein I-derived peptide library in patients with anti-beta 2-glycoprotein I antibody-associated autoimmunity. Hum. Immunol. 61, 366–377 (2000).
Eggenhuizen, P. J. et al. Smith-specific regulatory T cells halt the progression of lupus nephritis. Nat. Commun. 15, 899 (2024).
Greidinger, E. L. et al. T cell immunity in connective tissue disease patients targets the RNA binding domain of the U1-70kDa small nuclear ribonucleoprotein. J. Immunol. 169, 3429–3437 (2002).
Lu, L., Kaliyaperumal, A., Boumpas, D. T. & Datta, S. K. Major peptide autoepitopes for nucleosome-specific T cells of human lupus. J. Clin. Invest 104, 345–355 (1999).
Kanjana, K. et al. Autoimmunity to synovial extracellular matrix proteins in patients with postinfectious Lyme arthritis. J Clin Invest 133, https://doi.org/10.1172/jci161170 (2023).
Mohme, M. et al. HLA-DR15-derived self-peptides are involved in increased autologous T cell proliferation in multiple sclerosis. Brain 136, 1783–1798 (2013).
Lee, M. N. & Meyerson, M. Antigen identification for HLA class I- and HLA class II-restricted T cell receptors using cytokine-capturing antigen-presenting cells. Sci Immunol 6, https://doi.org/10.1126/sciimmunol.abf4001 (2021).
Joshi, N., Usuku, K. & Hauser, S. L. The T-cell response to myelin basic protein in familial multiple sclerosis: diversity of fine specificity, restricting elements, and T-cell receptor usage. Ann. Neurol. 34, 385–393 (1993).
Lolli, F. et al. Increased CD8+ T cell responses to apoptotic T cell-associated antigens in multiple sclerosis. J. Neuroinflammation 10, 94 (2013).
Fissolo, N. et al. Naturally presented peptides on major histocompatibility complex I and II molecules eluted from central nervous system of multiple sclerosis patients. Mol. Cell Proteom. 8, 2090–2101 (2009).
Yang, X. et al. Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides. Nature 612, 771–777 (2022).
Beltrami, A. et al. Citrullination-dependent differential presentation of a self-peptide by HLA-B27 subtypes. J. Biol. Chem. 283, 27189–27199 (2008).
Arakawa, A. et al. Melanocyte antigen triggers autoimmunity in human psoriasis. J. Exp. Med 212, 2203–2212 (2015).
Shen, Z., Wang, G., Fan, J. Y., Li, W. & Liu, Y. F. HLA DR B1*04, *07-restricted epitopes on Keratin 17 for autoreactive T cells in psoriasis. J. Dermatol Sci. 38, 25–39 (2005).
Lande, R. et al. The antimicrobial peptide LL37 is a T-cell autoantigen in psoriasis. Nat. Commun. 5, 5621 (2014).
Tiniakou, E. et al. Definition of naturally processed peptides reveals convergent presentation of autoantigenic topoisomerase i epitopes in scleroderma. Arthritis Rheumatol. 72, 1375–1384 (2020).
Bergseng, E. et al. Different binding motifs of the celiac disease-associated HLA molecules DQ2.5, DQ2.2, and DQ7.5 revealed by relative quantitative proteomics of endogenous peptide repertoires. Immunogenetics 67, 73–84 (2015).
Acknowledgements
We thank the members of the Joglekar laboratory, Dr. Xiaoxiao Wan, Dr. Cheryl Lichti, and Dr. Richard Jones for discussions regarding immunopeptidomics. This work was funded by NIH Director’s Transformative Research Award (1R01AT012826-01 to A.V.J.); NIH Director’s New Innovator Award (1DP2AI176138-01 to A.V.J.); and Breakthrough T1D strategic research agreement (3-SRA-2023-1354-S-B to A.V.J.).
Author information
Authors and Affiliations
Contributions
A.V.J. planned the direction of the review, edited and provided overall supervision. S.A. and B.C. conducted literature serach, wrote the original draft and edited the review. S.A. and B.C. are free to change the order in which their names appear.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Arshad, S., Cameron, B. & Joglekar, A.V. Immunopeptidomics for autoimmunity: unlocking the chamber of immune secrets. npj Syst Biol Appl 11, 10 (2025). https://doi.org/10.1038/s41540-024-00482-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41540-024-00482-x