Abstract
In modeling signal transduction networks, it is common to manually integrate experimental evidence through a process that involves trial and error constrained by domain knowledge. We implement a genetic algorithm-based workflow (boolmore) to streamline Boolean model refinement. Boolmore adjusts the functions of the model to enhance agreement with a corpus of curated perturbation-observation pairs. It leverages existing mechanistic knowledge to automatically limit the search space to biologically plausible models. We demonstrate boolmore’s effectiveness in a published plant signaling model that exemplifies the challenges of manual model construction and refinement. The refined models surpass the accuracy gain achieved over two years of manual revision and yield new, testable predictions. By automating the laborious task of model validation and refinement, this workflow is a step towards fast, fully automated, and reliable model construction.
Similar content being viewed by others
Introduction
Network-based dynamic modeling is an effective avenue toward understanding the response of biological systems to changes in their environment. The system is abstracted into an interaction graph (or interaction network), whose nodes represent the components of the system (e.g., proteins, cells, neurons, or species) and whose edges represent the directed, causal interactions among them. The dynamic model assigns each node a state variable and a regulatory function that determines the future state of the node given the current states of its regulators.
Boolean models are the simplest discrete dynamic models. They are used to model a variety of biological systems; examples include gene regulatory networks1, neuronal networks2,3, and ecological and social communities4,5. In Boolean models, the node state variables can take two values: 0, interpreted as low concentration or low activity, and 1, interpreted as high concentration or high activity. Boolean models are especially suitable for biomolecular networks due to the abundance of nonlinear, sigmoidal regulation in these networks6,7, and because of these models’ ability to describe perturbation (e.g., gene knockout) experiments. Through integrating the knowledge of the biology community, Boolean models successfully capture key behaviors in the biomolecular system of interest, and make useful predictions such as identifying master regulators or drug targets8,9 (see Supplementary Note 1 for examples). Predictions derived from Boolean models were verified experimentally in a variety of biological systems10,11. Multiple methodologies and tools can determine the possible long-term behaviors of Boolean models12,13,14,15,16. Here, we take advantage of minimal trap spaces to describe long-term behaviors. A minimal trap space, also called a quasiattractor, is a minimal set of states that the system can be “trapped” in and that can be characterized by fixing the values of some subset of the node state variables (see Supplementary Note 1 for a formal definition).
Due to recent advances in the analysis of Boolean dynamics12,13,14,17, the bottleneck in analyzing a biomolecular system is increasingly the time and effort involved in building the model. When high-throughput assays provide full state data (i.e., the state of all the relevant components at a given time), several viable automated Boolean model inference methods can be used18,19,20,21,22,23. For example, transcriptome data from multiple cell types can be used to infer gene regulatory network models for cell differentiation processes20,24,25. However, high-throughput assays are not the norm in cell signaling systems, which involve difficult-to-track post-translational modifications of proteins.
Traditional experiments, still frequently used in functional biology, measure a single component in two contexts, for example, in the presence or absence of a stimulus, or in the presence or absence of a perturbation of a different component. Compilations of such experimental perturbation–observation pairs are not equivalent to high-throughput measurements because certain components are more studied than others26 and because of inconsistencies between reported results27. The existing Boolean model inference methods are not suitable for such piece-wise, incomplete, and uneven data. In such systems, model construction is done via manual integration of distinct pieces of experimental evidence (see Supplementary Note 1 for more details of the modeling process). Although some parts of the model can be directly constrained by preexisting experiments, usually, many degrees of freedom remain. Modelers often use a process of trial and error informed by the insights of domain experts. Keeping the model up to date involves the same trial-and-error iteration. An illustrative example of the time and effort needed for model construction, validation, and update is the Boolean model of abscisic acid (ABA)-induced stomatal closure in the model plant Arabidopsis thaliana, which was introduced in 200628, significantly updated in 201729, and refined in 2018–202030,31.
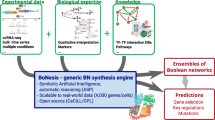
Our aim is to speed up and automate the trial-and-error process needed for model construction. Specifically, we consider the problem of refining and updating an existing baseline model to better agree with existing perturbation–observation data and also incorporate new data. We assume that the baseline model’s interaction graph describes the biological system relatively well, missing perhaps only a few edges. We develop a genetic algorithm-based workflow to adjust the Boolean functions of the model in a manner that optimizes the model’s agreement with curated perturbation-observation results. The workflow, and its implementation in the tool Boolean model refiner (boolmore), includes multiple ways to incorporate biological expertise that can limit the genetic algorithm’s search space to models that agree with biological knowledge. We demonstrate the effectiveness of our workflow by generating refined models of ABA-induced stomatal closure that agree significantly better with a compendium of published experimental results than the previous models.
Results
Outline of the boolmore tool
Our genetic algorithm-based workflow systematically tackles the huge number of modeling choices that must be considered during model validation and refinement. Genetic algorithms are a type of heuristic optimization that produces candidates through stochastic mutation and retains or eliminates them depending on a fitness score32. In the context of the problem considered here, the sought-after solution is the optimal refinement of an existing Boolean model, which consists of a signed interaction graph and the Boolean functions of each node. Boolmore uses the starting model to build a large number of mutated offspring models with different Boolean functions. These offspring models stay consistent with modeler-specified biological constraints and with the interaction graph, unless the addition of interactions is allowed. Boolmore scores the fitness of each model by comparing the model’s predictions to a compendium of experimental perturbation-observation pairs. Boolmore is meant to be used in conjunction with domain expertise to interpret the refined model and extract new predictions from it.
The previous works, most relevant to our method, involve genetic-algorithm-based inference of a Boolean model based on a directed network that integrates prior knowledge of interactions and regulatory relationships19,20,21,22,23,33,34. These algorithms take as input information a compendium of steady state values of all the nodes in the unperturbed system, complemented by steady state values obtained for perturbations. Another type of relevant prior work develops answer-set-programming methods to infer a Boolean model25 using reachability relationships between initial and final states (as available for cell differentiation) or to revise an existing Boolean model to better align with steady state or time course measurements35,36. We summarize in Supplementary Note 2 the goals and use cases of relevant previous algorithms. We provide a comparison of boolmore with the algorithms BoNesis25 and Gitsbe34 in our case study.
Here we provide a brief overview of the model refinement process of boolmore; see the “Methods” section for the details. Boolmore takes three different types of inputs: (i) the starting model, (ii) known biological mechanisms, and (iii) a categorized compilation of experiments. The starting interaction graph is derived from the starting model. The biological mechanisms are expressed as logical relations, such as “A is necessary for B.” The experimental results take the form of perturbation-observation pairs. Each pair describes the observed state of a node (biomolecule) in a certain context (e.g., in the presence of a signal, or in case of a knockout of another component). The observations are classified into five categories, including OFF, ON, and the intermediate category “Some”, which represents observations of an intermediate level of activation. We provide further interpretation of this category in Supplementary Note 3.
Boolmore repeatedly iterates through the steps depicted in Fig. 1. First, boolmore mutates the functions of the starting model to form new models, using a novel representation of monotonic Boolean functions. This representation has the advantage that it can easily constrain each mutation to preserve all the input biological information. For example, the new models stay consistent with the starting interaction graph, meaning that (i) the mutations preserve the sign of each edge, (ii) a mutation may delete a regulation represented by an edge of the starting interaction graph; this regulation can be later recovered, and (iii) a mutation cannot add a regulator, unless the user allows the addition of edges to the starting interaction graph from a user-provided pool. The representation also allows putting constraints on the mutation, such as preserving the “necessary” logical relation of certain regulators or locking a function to prevent it from mutating. Boolmore also generates crossover models in which each node’s regulatory function is randomly chosen from one of two models.
The starting model, known biological mechanisms, and a categorized compilation of perturbation-observation pairs are taken as input. These inputs are integrated directly into the model refinement process, as they are used to generate and score every single model. We introduced a novel representation to allow the mutation of Boolean functions while maintaining consistency with the interaction graph and the biological constraints. Boolmore generates model predictions using minimal trap spaces and employs hierarchy-based scoring to ensure that biologically realistic models receive higher scores.
Second, boolmore generates the predictions of the models by calculating the minimal trap spaces of each model under each setting, and identifying nodes that are ON, OFF, or oscillate. The averages of each node’s values over minimal trap spaces are taken as the model prediction. Third, boolmore computes the model’s fitness score by quantifying the agreement of the model predictions with the perturbation-observation results. To improve the alignment of biological relevance with scoring, the perturbations are grouped hierarchically, and the scoring of a perturbation-observation pair takes into account the perturbations at previous levels of the hierarchy. For example, for a model to get a nonzero score on the result of a double perturbation experiment, it must also agree with the observations of the constituent single perturbations. Finally, boolmore keeps the models with the top fitness scores, while also preferring models with fewer added edges. Boolmore contains multiple tunable parameters, including the mutation probability and the number of models generated in each step, which the users can freely customize. We present a parameter analysis in Supplementary Note 4.
Benchmark analysis
As is common practice in network inference, we first demonstrate the performance of boolmore through in-silico benchmark studies. We used 40 published Boolean models from the Cell collective repository37; this sample included all models with 30 or fewer variables. For each model, we generated artificial experiments, each consisting of a perturbation (fixing the state of a set of nodes) and the observation of the state of a different node. We used 80% of the artificial experiments as a training set to refine an initial model that had the same interaction graph as the actual model but its regulatory functions were randomly selected. The remaining 20% of the artificial experiments were used as a validation set to test the predictive power of the refined model. We describe the details of these benchmark studies in the “Methods”.
We found that the starting models had on average a 49% accuracy on the training set, and boolmore improved the models to 99% accuracy on average (Fig. 2). Notably, boolmore also increased the accuracy of the models on the validation set from 47% on average to 95% on average. This indicates that boolmore does not overfit the training set and that the refined models give valid predictions.
A Illustration of the accuracy improvement on the training set (80% of the artificial experiments) over the iterations of the algorithm. The blue circles represent the highest accuracy obtained at each iteration, averaged over 200 independent runs, with the error bars representing the standard deviation over the runs. B For each model, the average accuracy on the training set (80% of artificial experiments) of five starting model variants is shown in orange, and the average accuracy of the five refined models is shown in blue. Error bars represent the standard deviation across the five replicates. The names of each model and their accuracies are listed in Supplementary Table 1. Orange and blue dashed lines indicate the average accuracy of the starting models (49%) and the refined models (99%), respectively. C The average accuracies on the validation set (20% of artificial experiments) of the five starting model variants (orange) and the five refined models (blue). Boolmore had no information on the validation set in its model refinement process, but was still able to improve the accuracy greatly, from 47% on average to 95% on average.
Improving the ABA-induced stomatal closure model
To go beyond simple benchmarks and use our approach to search for new biological insights, we apply our method in a case study. We selected a system that is representative of the challenges inherent in constructing a Boolean model of a complex biological phenomenon and keeping it up-to-date with current literature. Specifically, we analyzed a group of Boolean models, each aiming to integrate the mechanisms through which the hormone ABA leads to the closure of plant stomata. The biological details of this process are provided in Supplementary Note 5. The first model of ABA-induced closure, published in 2006 by Li et al.28, included 42 nodes. This was expanded to 81 nodes in 2017 by Albert et al.29. An alternative expansion to 60 nodes was published in 2018 by Waidyarathne and Samarasinghe38. The 2017 model was refined in 2019 by the addition of a new edge and a simultaneous attractor-preserving reduction to 49 nodes30. We decided to use the 2017 model (whose node names are given in Supplementary Table 2) as the basis of model refinement, as it is the most comprehensive and its analysis included a thorough comparison with 112 perturbation-observation pairs. The reported accuracy of the 2017 model was 95/112 = 85%.
Despite its overall high accuracy, the 2017 model also exhibited two important weaknesses. First, 13 nodes were shown experimentally to lead to closure when perturbed by external interventions. The 2017 model failed to recapitulate nine of these observations according to the simulation-based criteria used at the time. Second, plant stomata reopen after the removal of the ABA signal, enabling the plant to resume photosynthesis. This reversibility of stomatal closure is not captured by the model.
Two follow-up publications aimed to address each of these two weaknesses of the 2017 model by making parsimonious hypotheses. Maheshwari et al.30 hypothesized in 2019 and experimentally confirmed the existence of an additional edge, thereby recapitulating five of the nine experimental observations of closure that were not captured by the 2017 model. A second follow-up to the model in 202031 identified that the source of the irreversibility of stomatal closure in the model is the assumption of self-sustained activity of four nodes. The 2020 model achieved reversible closure and preserved the 2019 model’s success in capturing the five experimental observations of closure. We describe the 2017 model and its two follow-ups in more detail in Supplementary Note 5. We emphasize that both of these improvements were proposed after an in-depth analysis of the 2017 model’s weaknesses and individual exploration of many hypotheses to address these weaknesses while preserving the strengths of the model.
We aimed to refine the 2017 model such that it reproduces the interventions that lead to stomatal closure and exhibits reversible closure. Importantly, we aimed to do this without including these criteria as explicit constraints of the refinement algorithm. We undertook an extensive search of the experimental literature and expanded the number of perturbation–observation pairs to 505 (listed in Supplementary Table 3). Noting that the 2017 model had an erroneous edge, recognized in a later publication39, we considered two starting models, baseline A and baseline B, which correct the model in two different ways. The key difference between the two models is that in the presence of ABA, the baseline model A features the oscillation of multiple nodes, driven by transients in the elevation of the cytosolic Ca2+ level, while baseline model B defines an abstract node “Ca2+c osc” and leads to a fixed state for all nodes. Supplementary Note 6 provides a more detailed description of the two baseline models. In our application of boolmore on either baseline model, we adopted specific constraints for the regulatory functions of 26 nodes and specific criteria for allowing 13 additional, experimentally-supported edges, as we describe in Supplementary Note 7. The algorithm added 8 edges from the pool, which we describe later as new predictions.
It took roughly 10 h to go through 100 iterations of the algorithm and generate 10,000 models on a PC with an AMD Ryzen 5 3600 6-Core CPU at 3.8 GHz. The majority of the computation time was spent on model evaluation; the evaluation of each model took roughly 7 s on average.
The application of boolmore significantly improved both baseline models. We will refer to the model obtained after applying boolmore to baseline model A as genetic-algorithm (GA)1-A and the model obtained after refining baseline model B as GA1-B. We indicate the regulatory functions of the GA1-A and GA1-B models in Supplementary Note 8. Our first specific model refinement goal was to reach better agreement with the experimental interventions that cause closure in the absence of ABA but were not recapitulated by the 2017 model.
Better agreement with experimental interventions that yield closure
Table 1 summarizes these experimental observations and the model results for the node Closure. The categorization of the experimental results, ranging from Some to ON, reflects the observed degree of closure (decrease of the stomatal aperture) induced by each intervention. The table indicates the results of the 2017 model, the Maheshwari et al. 2019 model, and the two GA-refined models. The GA1-A model preserves the 2017 model’s agreements and has an improved score for three additional responses. The GA1-B model preserves the agreement for three interventions, has a lower score than the 2017 model in one case (supplying InsP3), and receives a higher score for three additional interventions. Note that the elements whose closure-inducing nature is newly recapitulated by the GA-refined models lie at the core of the system, as reflected in the large number of experiments studying them (see the last row of Table 1).
Reversibility
The 2017 model has 17 minimal trap spaces in the absence of ABA; 16 with Closure = 0 and 1 with Closure = 1. In the presence of ABA, the model has a single minimal trap space with Closure = 1. If the model starts in this trap space, and then ABA is taken away, the model can only reach the single minimal trap space with Closure = 1, and therefore fails to achieve reversibility. The elimination of this trap space would yield a one-to-one correspondence between the signal ABA and the closure response. Boolmore’s scoring method prefers models whose minimal trap spaces are completely aligned with the experimental observations. Specifically, when an experimental observation corresponds to a lack of closure (Closure = 0) but the model prediction (i.e., the average value of the node Closure in the minimal trap spaces) is close to but not equal to zero, the model receives a partial score. Both GA1-A and GA1-B succeeded in eliminating the trap space with Closure = 1 in the absence of ABA and thus achieved reversibility. It is particularly remarkable that this reversibility was achieved without the introduction of time-dependent regulatory functions as done in the 2020 revision31. The score gain from achieving reversibility is reported in Table 2. Note that the GA-refined models were not able to gain the maximum score improvement from reversibility due to trade-offs in capturing some of the experiments (i.e., due to the fact that achieving agreement with one experiment may create a disagreement with another experiment).
Better agreement with experimental results
As 1 point in the score of a model means recapitulating one perturbation-observation pair, the highest possible score for a GA-refined model is 505. The 2017 model recapitulated 33 pairs with a score of 1, had a score of 0.9 for 257 pairs, a score in the range 0.5–0.8 for 50 pairs, and a score of 0.4 or lower for 165 pairs, achieving a score of 310.5/505 (61.5% accuracy). An improved model would need to preserve the original model’s agreements with the experimental results and reach agreement with the experiments that the original model did not capture. The score of GA1-A is 84.5%, significantly improved compared to the accuracy of 61.7% of the baseline model A, which is closest to the 2017 model. The accuracy of GA1-B is 80.8%, a dramatic improvement compared to the accuracy of 36.5% of the baseline model B, which uses an abstract node “Ca2+c osc.” We confirmed that the increase in the score of both GA1-A and GA1-B is due to resolving the majority of the original model’s discrepancies with experiments (see Table 2).
To place these results into context, we present the accuracy of the previous manually constructed models as well as two additional versions of GA-refined models in Fig. 3. In the GA0 models, we did not allow the addition of edges, therefore the GA0 models stay consistent with the original interaction graph. In contrast, for GA2 models, we allowed more assumed edges, without requiring experimental support. We describe these models in detail in Supplementary Note 10. The figure indicates that all versions of GA-refined models surpass all published models, while at the same time decreasing the time needed for model refinement.
The accuracy of the models marked with * is scaled to the percentage (50–75%) of experiments that apply to them. We note that 100% accuracy is not possible due to intrinsic limits on the agreement between the experiments on ABA-induced closure and Boolean models. We discuss these limits in Supplementary Note 9. The GA0 and GA2 models are variants of GA1 in terms of the allowed addition of edges.
We evaluated the reproducibility of the genetic algorithm-based refinement by performing 16 independent GA1 runs starting from the baseline A model, which is closest to the 2017 model. The mean of the final accuracies was 81.5% with a standard deviation of 2.1%. The GA1-A model had the highest final accuracy, 84.5%. There was a significant consensus among the GA1 model variants (e.g., 16 of the 35 functions that could change converged into an identical or logically equivalent form) as well as subtle dissimilarities that explain the differences in the score. The fitness score saturated before 100 iterations in each run. This shows that although Boolmore was not able to find the global maximum in every run, it was able to converge on a local maximum. This is a remarkable result considering the extremely high number of possible models for this case study. Indeed, we calculated using Dedekind numbers that the rough lower bound of the number of Boolean models consistent with the interaction graph and the constraints is 10153.
The goal of our model refinement workflow was to find biologically relevant improvements to an existing model, mirroring a manually refined model. Comparison with two publicly available tools for Boolean model inference or refinement, BoNesis25 and Gitsbe34, indicates that neither tool is able to achieve this goal for the ABA-induced closure process. BoNesis could not return a single model within 24 h even when restricting the experimental input to the perturbation-observation pairs satisfied by a baseline model. All the models generated by Gitsbe in over 13 h contained many biologically invalid modifications to the regulatory functions, as Gitsbe lacks the ability to constrain the functions to preserve biological mechanisms. Even if overlooking the lack of biological interpretability, all of the Gitsbe-generated models had a lower score than the worst-scoring boolmore-refined models. See Supplementary Note 11 for a detailed description of our comparison process.
The model refinements identified by boolmore have both explanatory and predictive power. The existence of two alternative baseline models allows the identification of changes implemented by boolmore in both models. These changes, which are listed and interpreted in Supplementary Note 12, have a high likelihood of being biologically meaningful. In addition to an increased understanding of the biological system, it is also possible to extract novel predictions from the GA-refined models and suggest new experiments. In the following, we describe a selection of new predictions.
New predictions of the GA-refined models of ABA-induced closure
Modifications to the interaction graph
Both GA-refined models feature significant changes to the interaction graph, which can serve as new predictions. GA1-A deleted 24 edges out of 152 starting edges. GA1-B deleted 17 edges out of 145 starting edges. Ten edges were deleted in both models; these edges are shown with red dashed lines in Fig. 4. A significant fraction of these edges represented assumptions of the 2017 model based on indirect evidence. These assumptions are no longer needed due to the improvements to the regulatory functions made possible by boolmore (see Supplementary Note 12 for examples). In some of these cases, we could identify a shortcoming in the reasoning that led to the unnecessary inclusion of a variable into a regulatory function in the original model. These findings illustrate how an automated method using a genetic algorithm can overcome modelers’ bias in selecting edges and functions, revealing optimal possibilities. Note that the deletion of an edge does not necessarily mean lack of influence; the influence may be preserved through a path, or the deletion may indicate that in the context considered the influence is not significant enough to overcome the effects of the other regulators in a phenotypically relevant way.
Each edge that terminates in an arrowhead indicates a positive regulation, and a round tip means negative regulation. Blue edges are shared additions of the GA-refined models, and red dashed edges were deleted by both GA-refined models. The nodes with a yellow background are the key intervention nodes whose perturbation leads to some degree of closure. The full names of the elements are indicated in Supplementary Table 2.
GA1-A added six new edges, and GA1-B added seven new edges from the pool of 13 experimentally supported new edges. The added edges present in both GA-refined models indicate biological mechanisms that were not included in the 2017 model. As shown in Table 3, five new edges were added in both models. Importantly, the added inhibitory edge between PA and ABI2 recapitulates the experimentally supported prediction of the 2019 follow-up to the 2017 model30. The success of these shared additions confirms the improvements possible from the incorporation of new biological information.
Predictions that can be verified experimentally
The GA-refined regulatory functions also serve as new biological predictions. We summarize selected testable predictions in Table 4 and explain them in Supplementary Note 13. Here we illustrate the types of predictions with a few examples.
The GA-refined models modify the regulatory function of the anion channel SLAC1 such that it is easier to activate. As a consequence, the models recapitulate the experimental observation that ROS activates SLAC1. A follow-up prediction is that the resulting anion flow brings the malate concentration below the threshold.
Causal relationships mediated by chains of interactions (pathways) can also yield new predictions. The 2017 model and the GA-refined models agree in predicting that ROS is sufficient to induce PA production. This is experimentally testable. As the GA-refined models incorporate the new observation that PA is sufficient for microtubule depolymerization, a follow-up testable prediction is that ROS can induce microtubule depolymerization; this can be tested by methods used by Eisinger et al.40.
The shared features of the GA-refined models’ minimal trap spaces, which are described in detail in Supplementary Note 14, identify further predictions. One such prediction is that external Ca2+ would yield no or a very limited amount of ROS production. While ROS production in ABA-induced closure has been experimentally documented, the production of ROS in response to external Ca2+ has not yet been studied experimentally.
A more general insight can be gained from observing that GA1-A and GA1-B rely on two different mechanisms to yield minimal trap space results of 0.5 that achieve agreement with experimental observations classified as “Some” in Table 1. The attractors of GA1-A feature oscillations in Closure along with a significant number of other nodes, which are due to the oscillations of Ca2+c. In contrast, GA1-B has two attractors, one featuring Closure = 1 and the other featuring Closure = 0. Although different in a technical sense, these results are consistent with each other in that they both suggest population-level heterogeneity of the stomatal responses. Due to the challenges of tracking individual stomata in real time, there are few studies of individual stomata. Nevertheless, the studies that exist include observations of multiple types of oscillations in the stomatal aperture, including Ca2+ −induced oscillations (reviewed by Yang et al.41). In addition, Li et al.28 reported a significant bimodality of the ABA-treated stomatal aperture distribution. Our results suggest that more in-depth analyses of the time-dependent status of signaling mediators in individual guard cells may reveal a richer dynamic picture than previously thought.
Discussion
Small, local changes in a Boolean model, such as to the regulatory function of a single gene, can lead to global changes in the model’s attractor repertoire and thus to the predicted phenotypes. This makes the process of iteratively building a model, or incorporating new biological knowledge into an existing model, extremely challenging. Here we automate the process of refining or updating an existing Boolean model and implement it as the tool boolmore. We use a genetic algorithm to adjust the regulatory functions of the model to improve its agreement with curated experimental results. Automated exploration and quantitative scoring of modeling decisions can alleviate the immense cognitive burden of integrating myriad experiments into a causal model, while reducing human error and modeler bias. Our workflow allows modelers to automatically explore many more modeling choices than previously possible, enabling systematic evaluation of alternate modeling assumptions with large, global effects on the model dynamics. As an illustrative example, we considered a model of ABA-induced closure (baseline model B) in which the oscillating negative feedback between cytosolic calcium and calcium ATPase was replaced by a single node whose activity indicates calcium oscillation. Using boolmore, we refined baseline model B, which had an accuracy of only 36.5%, into model GA1-B, with an accuracy of 80.8%.
Any method that involves fitting to data has a chance of overfitting, i.e., inferring more parameters than can be justified by the data42. The parameters of a Boolean model are the Boolean functions of individual nodes. In our case study, boolmore deleted many edges that were included in the manually constructed starting models. Deletion of edges is analogous to removing parameters, and hence, boolmore actually helped reduce the risk of overfitting that exists in the manual refinement process. Our workflow also helps introduce new edges in a much more conservative manner by only allowing edges that increase the accuracy of the model as a whole. The mutation of the regulatory functions used in boolmore preserves these functions’ biological interpretability. The representation of the functions preserves the sign of each regulatory relationship. The constraints ensure that known mechanisms are reflected in every model version. The predictive power of the models refined by boolmore is evident in the benchmarks, where the refined models showed 95% accuracy over the validation set, which was not used in the refinement process.
Boolmore uses a flexible scoring method that appropriately handles low-throughput state data and the subtleties that arise in discretizing experimental data. In the case study considered here, for example, we used this flexibility to introduce additional outcome categories (e.g., “Some”, “OFF/Some”) to describe intermediate outcomes (e.g., reduced closure) and inconsistency between repeat experimental observations. By considering the interdependence of perturbation experiments, our scoring achieves an unprecedented level of biological realism even with limited data.
The scoring system can be readily augmented by additional measures, such as the number of attractors or the phenotype transitions under different environments, to produce more realistic models. Model prioritization can be customized so that only models with certain key behaviors are selected. Furthermore, the modeler has the freedom to determine how individual experimental observations should be encoded; for instance, allowing for categories, such as “Oscillating” or “Bistable” (see Supplementary Note 3 for the types of experiments that lend themselves to this categorization).
Although boolmore was implemented for locally-monotonic Boolean functions, it can readily handle non-monotonic Boolean functions as well as multi-level variables. Biologically justified context-dependent regulation (expressed as a non-monotonic function) can be incorporated via virtual mediators (see Methods). As a lossless mapping between a multi-level variable and a set of Boolean variables has been worked out43,44, boolmore can accommodate multiple levels. We indicate in Supplementary Note 15 the details of this adaptation and a proof of concept application to a model of nutritional regulation of lisosomal lipases in C. elegans45. A multi-level representation of the output variable “Closure” is especially needed in future models that integrate the response to the various signals that lead to various degrees of stomatal closure. These signals include ABA, high CO2, darkness, and their combinations. Boolmore can be a useful tool for identifying the number of levels that yield the best agreement with experiments.
Boolmore needs a high-quality interaction graph. Our case study indicates that even one erroneous edge can significantly limit the success of model refinement (see Supplementary Note 10 for related results on our case study). Fortunately, many methods have been developed to construct or infer biomolecular interaction graphs46,47, and curated interaction graphs are available in multiple databases48,49,50, increasing the likelihood of a high-quality interaction graph for any cellular process to be modeled. Gaps of knowledge in the interaction graph can be filled by allowing the addition of edges, increasing the accuracy of the model (see Supplementary Note 10 for related results). Each such addition serves as a new prediction. However, edge addition is only effective in moderation, as each additional edge doubles the size of the search space.
Model evaluation is the major computational bottleneck of boolmore and we believe that no trivial speedup is possible. By relying on the Python package pyboolnet13, boolmore could find the minimal trap spaces in our 82-node case study in a hundredth of a second. Even with this remarkable speed, evaluating more than 8000 models under more than 260 perturbation settings resulted in 10 h of runtime for a single run on a personal computer.
Although the fast computation of minimal trap spaces is a major contributor to boolmore’s effectiveness, the focus on trap spaces also imposes some limitations. Boolmore cannot incorporate initial states, timecourses, and cannot determine basins of attraction. Consequently, boolmore cannot test whether a model can reproduce an organized set of global states corresponding to an oscillation, such as the cell cycle or a circadian oscillation. In some cases, timecourse information can be abstracted into early and late events, using a suitable categorization of interactions51. When comparing to experiments describing early responses, boolmore would mask (inactivate) the late-event interactions, and use the trap spaces of the model version that only has the early events. Other limitations can be overcome by integration of boolmore with a simulation tool, e.g., using the fast GPU-based simulator cubewalkers52. Such integration would allow identifying the long-term behavior specific to a certain initial condition and determining the relative basin of each minimal trap space. In turn, this information could be incorporated into scoring schemes to evaluate model agreement with experimental observations on cell subpopulations. Integration with a simulator would also allow the probabilistic implementation of intermediate perturbations (knockdowns) and open the way toward the incorporation of probabilistic Boolean models53,54.
Our automation of model refinement streamlines model-building by lowering the hurdle for the initial model. Once the experimental database and an interaction graph for the model are set, a quickly achievable preliminary model can be refined in a fraction of the time that would be needed for manual model building. We envision that boolmore may also be used as an exploratory tool, allowing biologists and modelers to evaluate whether, and how, a model can accommodate hypothetical experimental results. Boolmore can also be integrated with other steps, such as automated evidence gathering55 and model expansion56 to progress toward fully automated and extremely fast model construction.
Methods
Methodological details of boolmore
Mutating functions such that edge signs are preserved
The interactions and regulatory relationships in biological networks are locally monotonic in the vast majority of cases. This means a regulator either inhibits or activates its target; it is not an inhibitor in one context (i.e., for a certain state of other regulators of the target node) and an activator in another context. The signs of the interactions are built from the literature, and form the foundation of the model. These interactions are often well established, and hence changing the signs will lead to completely unrealistic networks. Boolmore only allows Boolean function mutations that preserve the original signs. Boolean models of biological networks may contain functions with context-dependent regulation; indeed, 1% of the functions of an ensemble of 122 Boolean models were found to be context-dependent57. Such non-monotonic functions can be handled by introducing virtual mediators, as we describe later.
To achieve random mutations of Boolean functions that preserve the original signs, we propose a degenerate binary representation of each function based on a disjunctive normal form of the function. The key idea is the following: a function with p positive regulators and n negative regulators can be expressed (not necessarily uniquely) as the disjunction (OR composition) of a subset of the 2n+p conjunctions (AND compositions) consistent with the regulatory signs. Each of these conjunctions is assigned a location in a binary string of length 2n+p; the binary string is interpreted as the disjunction of the conjunctions corresponding to the locations in which the string has a 1. Each mutation changes a randomly selected digit of this binary representation and is guaranteed to preserve the signs of the regulatory relationships.
The specific representation for a positive regulatory function (i.e., one for which all regulators are activators) with k inputs is
Here, Xi represents the state of the ith input node out of k input nodes. The notation “|” means logical “OR” and “&” indicates logical “AND”. A constant Boolean coefficient aS is assigned to each subset S of input nodes, from S = {} to S = {1,…,k}. Note that this representation is not unique. The coefficients aS can be ordered to obtain a binary representation of the function. A natural ordering interprets each S as the binary representation of the numbers from 0 to 2k–1. This ordering is also traditionally used in the truth table representation of Boolean functions.
For example, let us consider three variables A, B, C, and the Boolean function f(A,B,C) = B&C|A&B. This function can be written as f(A,B,C)=0|0&C|0&B|1&B&C|0&A|0&A&C|1&A&B|0&A&B&C. Note that this representation includes all the possible subsets of ABC, leading to 23 = 8 clauses; the original three clauses are the terms that start with 1. The Boolean function can be represented by a string of the aS coefficients, i.e., f(A,B,C) is represented by 00010010. The benefits of this representation are that any possible combination of 0s and 1s represents a positive (activators-only) function, and that the combinations span all the positive functions. Exemplifying the degenerate nature of this representation, 00010011 also represents f because A&B&C is implied by B&C|A&B. However, the representation that has the maximal number of 1s (max representation) is unique and is equivalent to the truth table of the function (00010011 in this example). The representation that has the minimal number of 1s (min representation) is also unique and is equivalent to the Blake canonical form of the function (00010010 in this example).
To obtain a mutated function, each digit of this representation is changed with a certain probability. For example, the mutation may change the sixth digit and lead to 00010110. The mutated function is thus f’(A,B,C)=0|0&C|0&B|1&B&C|0&A|1&A&C|1&A&B|0&A&B&C, which can be further simplified to f’(A,B,C) = B&C|A&C|A&B.
This method can be extended to mutate locally monotonic functions that do contain the NOT operator (which we will represent as !). If g(A,B) = !A&B, we can use the change of variables A’ = !A and represent the function as the positive function g’(A’,B) = A’&B. As long as we keep the original records of the signs, any locally monotonic function can be switched to a positive function, mutated, and switched back.
Note that the above representation naturally has a bias toward 1. For example, if the first digit of the binary representation is mutated to 1, the whole function becomes 1. To remove this bias, we also consider a binary representation of the negation of the function. For example, the negation of the above function, !f(A,B,C) = !B | !A&!C = !f(A’,B’,C’)=B’|A’&C’ can be represented as 0010100. In the negated representation, mutating the first digit to 1 makes the negated function 1, and thereby makes the original function 0. To remove bias toward one output state or the other, we introduce a 50% chance to mutate the negation of the function rather than the function itself.
Although boolmore’s encoding of Boolean functions relies on their local monotonicity, it can flexibly handle non-monotonic functions. Modelers should simply introduce virtual mediators for each regulator whose effect is non-monotonic. This allows the function to be fixed or mutated depending on the choice of the modeler. For example, if a variable C is governed by the function “A XOR B”, the modeler can add mediators a and b, and transform the system into
\(\begin{array}{ll}{{\bf{a}}}^{\ast }={\rm{A}}\\ {{\bf{b}}}^{\ast }={\rm{B}}\\ {{\rm{C}}}^{\ast }=({\rm{A}}\,\mathrm{or}\,{\rm{B}})\,\mathrm{and}\,\mathrm{not}\,({\bf{a}}\,\mathrm{and}\,{\bf{b}})\end{array}\)
In cases where there is good biological justification for the XOR function, the modeler can include nodes a, b, and C in the group of nodes whose Boolean function is fixed. Depending on the situation, the modeler could choose to let the function of C to be mutated, to allow testing models with various modifications of selected parts. For example, if one is more certain about the positive effects of A and B, then A and B could be added to the list of required regulators, while the negative effects represented by a and b are allowed to be dropped.
Preserving known mechanisms via constraints
In some cases, the biochemical mechanisms of certain regulatory relationships are known. We encode such knowledge as constraints to the Boolean regulatory functions, in a similar vein as Azpeitia et al.58. These constraints not only ensure reasonable models, but they also reduce the search space greatly. For example, if it is known that the activation of a regulator A is necessary for the activation of the target B, then the form of the Boolean function of B is constrained to be “fB=A & (other regulators)”. For a function with four inputs, this reduces the number of possible Boolean functions with fixed signs from 168 to 20. The constraints of the regulatory functions are enforced by their binary representations. For example, if A is constrained to be a necessary regulator, any term that does not contain A will have a coefficient of 0 in the binary representation.
We implemented five types of constraints in our case study. Four explicit constraints are fixed functions (not allowing the mutation of a function that describes a known mechanism), edge preservation (not allowing the loss of a biologically well-supported edge), logic preservation (preserving the information that a regulator is necessary for the activation of the target node), and grouped regulation (preserving the relationships that express an enzyme-catalyzed reaction). The fifth constraint is that, in general, we do not allow mutations that would transform a node into a source node (e.g., by the loss of its last remaining regulator). Exceptions from this constraint are explicitly listed.
The currently implemented constraints are based on our current experience with using boolmore on a signal transduction network. Future applications will likely reveal new constraints, which can be implemented.
Allowing the addition of new edges from a limited pool of experimentally-supported hypotheses
Boolmore modifies the interaction graph by deleting or adding edges. The deletion of edges is done implicitly through the modification of regulatory functions. The addition of edges was implemented in a restrictive way: boolmore can only select edges from a predetermined pool. Initially, a newly added regulator is integrated with the rest in a random way, and the function can mutate through the iterations. For example, consider that node X, which had the original function fX(A,B,C) = B&C|A&B|A&B&C, acquires a potential new positive regulator D. Its binary representation now has 16 digits instead of eight, thus acquiring 8 free parameters, which we mark with the symbol “?”: fX(A,B,C,D)=0|?&D|0&C|?&C&D|0&B|?&B&D|1&B&C|?&B&C&D|0&A|?&A&D|0&A&C|?&A&C&D|1&A&B|?&A&B&D|1&A&B&C|?&B&C&D=0?0?0?1?0?0?1?1?. If each “?” is set to zero, then the additional regulator is fully redundant, whereas the regulator is sufficient for activation if the first “?” is equal to one. When adding an additional regulator in boolmore, we initialize each “?” randomly, with a 50% chance to be 0 or 1.
Models are given internal penalties when adding edges, so that models in which the addition of an edge did not lead to a score increase are less likely to survive through the iterations. This is done by counting the number of added edges and prioritizing the model with fewer added edges among two models with the same score. We also prioritized models with simpler functional forms. This was done by counting the number prime implicants (or equivalently the number of 1s in the minimal binary representation) and prioritizing the model with a smaller number whenever there are two models with the same score and the same number of added edges. This method helps prevent the models from deviating too much from the original interaction graph and from increasing their complexity. However, even with these preventive measures, each edge in the pool makes the search space exponentially larger, and can preclude boolmore from finding an optimal model in a reasonable amount of time. Hence we only allowed the addition of user-provided edges, often limited to edges with experimental support.
Interpreting experimental results in a Boolean context
Boolmore computes each model’s fitness score using input data consisting of experimental perturbations and a coarse-graining of the observed outcomes. We classified the experimental observations into five categories: OFF, OFF/Some, Some, Some/ON, ON; we describe below how we assigned these categories for each node, though we note that our workflow is flexible and allows other choices.
Experimental perturbations, such as knocking out a gene or providing excess amounts of a protein have natural Boolean interpretations; the corresponding nodes are considered to be fixed OFF for the former and ON for the latter. The observations of mRNA or protein concentrations in the unperturbed (wild type) or perturbed (e.g., mutant type) systems have a continuous spectrum of outcome. We use a comparative method to express these observations in a form that is compatible with Boolean dynamics. In the case of a signal transduction network, we use the observed concentration or activation of each node in the presence/absence of the signal (in the wild type system) as two points of reference, akin to a positive and negative experimental control. We coarse-grain the observed node activities in response to perturbations by comparison to these two points of reference.
We illustrate in Fig. 5 the case of a node R, which has higher activation when the signal S is present in the wild type. We consider this level of activation to be the ON state (R = 1), and in any perturbation that leads to a similar activation level or higher, R is considered ON. The levels are assigned to the OFF state in a similar manner. In addition to the preferred OFF and ON categories, we also introduced intermediate and mixed categories as necessary. If the observed activation level is an intermediate between the OFF and ON states, it is categorized as “Some”. Although a Boolean model has no simple way of describing such an intermediate level, it can be realized by an attractor in which the node oscillates or by multiple attractors, with the node being with ON in some of the attractors and OFF in the others. We discuss the interpretation of the “Some” category and its possible customization in Supplementary Note 3. We assigned the OFF/Some or Some/ON categories in cases when there are multiple reported observations for the same perturbations that have non-identical results, or in cases where a clear comparison with the reference was impossible.
A The reported results are normalized by the activation of the response node R in the absence of the signal (S = 0). The wild type shows higher activation of R in the presence of the signal S. M and C represent mediators of the signal. ”KO” means knockout and “CA” means constitutive activation. The wild type response under S = 0 is categorized as OFF; the wild type response under S = 1 is categorized as ON. If the value of R observed under a perturbation is similar to one of these two reference values, it will be included in the same category as the reference. For example, the response to [S = 1, C CA] is categorized as ON. Notably, the response to [S = 0, C CA] is categorized as Some. B Illustrative example of determining the minimal trap space agreement and the hierarchy-based score of putative models. Each model indicates the next state of R (denoted R*) as a function of the current state of S or M. The model with R* = 0 has the same minimal trap space agreement as the R* = S model, and in particular it agrees with the observation [S = 1, M KO] → R = 0, but it receives a lower hierarchy-based score because of its discrepancy with the observation [S = 1] → R = 1. In general, the difference between the minimal trap space agreement and the hierarchy-based score (highlighted in yellow) is more prominent in more complex perturbation experiments.
Detailed description of the model’s outcome
We use pyboolnet, a Python package for analyzing Boolean models13 to determine the minimal trap spaces of the model under various constraints that mimic perturbation experiments. Minimal trap spaces are a close, update-scheme-independent approximation of attractors and their identification is more computationally efficient (see Supplementary Note 1). For each minimal trap space, nodes constrained to be ON are assigned the value of 1, nodes constrained to be OFF are assigned the value of 0, and unconstrained (oscillating) nodes are assigned the value of 0.5. In each comparison with the experimental observation obtained in a perturbation condition, the model outcome is the average value of the observed node in the minimal trap spaces corresponding to that condition. For example, if there are two minimal trap spaces and the node oscillates in one and has the state 1 in the other, the average node value is 0.75.
Although there are alternative tools for minimal trap space calculation that can outperform pyboolnet in typical settings, it is optimal for boolmore. This is because pyboolnet allows very fast computation of minimal trap spaces using the Blake canonical form of the functions, and its main bottleneck is calculating the Blake canonical form from the given functions. In boolmore, the binary representation allows the computation of the Blake canonical form at a very small cost.
Scoring model fitness with a hierarchy-based method
Each model receives one point toward its fitness score per recapitulated perturbation-observation pair. To ensure that the simulated perturbation is causally linked to the model outcome, a perturbation experiment is considered to be recapitulated only if the model predictions agree with measurements obtained for subsets of the perturbation as well, resulting in a hierarchy of experimental observations. The top of the hierarchy is the ‘resting state’ observation of the wild-type system in the absence of any signal. The signal-response pairs of the unperturbed (wild type) system are one step down, as are the observed responses to perturbations of single intermediary nodes in the absence of any signal. The signal-response pairs under perturbations of single mediators are two steps from the top. Perturbations of multiple mediators are at increasingly lower levels of the hierarchy.
As an illustrative example, consider a system in which a signal S leads to a response R through a mediator M (see Fig. 5B). The knockout of the mediator M (M KO) inhibits the response to the signal (R = 0 even if S = 1). A hypothetical model that says R is inactive regardless of the signal (R* = 0) recapitulates the result that M KO leads to R = 0. However, this model does not respond to the signal even when M is not knocked out. Our scoring method ensures that a high-scoring model satisfies the top-of-the-hierarchy experiment [S = 0] → R = 0, the one step down experiment [S = 1] → R = 1, as well as the more complex experiment [S = 1, M KO] → R = 0. Note that S = 0 is the default and should be included in the specification of the experiment unless S = 1.
Boolmore scores each model in a two-step process. First, it determines the agreement of the model with the observation of each perturbation experiment, and then it scores the model by considering all agreements with the experiments at higher levels of the hierarchy. For each perturbation condition, the model prediction is the average value of the observed node in the minimal trap spaces. The model’s agreement for that perturbation experiment is given depending on how well the prediction agrees with the categorization of the experimental observation. Each agreement function’s output ranges from 0 to 1 following a piece-wise linear mapping, indicated in Table 5. For example, if the experimental outcome is categorized as ON, a model with a prediction of 1 receives an agreement of 1 for that experiment and another model with a prediction of 0.5 gets an agreement of 0.5. The final score is the product of all the attractor agreements of the subset perturbation experiments. For example, in Fig. 5B, if we are considering the perturbation [S = 1, M KO], the score is given by multiplying the agreements of 4 experiments, namely [S = 0], [S = 1], [S = 0, M KO], and [S = 1, M KO] itself.
Setups used for the benchmarks and case study
Parameters for running the genetic algorithm
The steps described in Fig. 1 are performed on a population of 100 models generated in each iteration. We do 100 iterations in each run. The top 20 models with the highest fitness scores from the previous iteration are carried over. We perform 20 repetitions of selecting two models randomly from the top 20 (fitter models having a higher chance of being selected) and generating a cross-over model in which each node’s Boolean function is chosen with equal probability from one of the two models. This process generates 20 additional models. Finally, from this pool of 40 models, we repeat 80 times the process of selecting a model and mutating it, to generate the remaining 80 models of the new iteration. We used a mutation probability of 0.01 in the functions. Fitter models have a higher chance of being selected in this process as well. Ten thousand networks are generated in a single run. These numbers were chosen such that the best score saturates by the end of the run in the benchmarks and the case study. We performed a parameter analysis and found significant robustness to changes in these parameter values (see Supplementary Note 4 for more details).
We found that the performance of the genetic algorithm does not depend sensitively on the choice of the parameters. The most important choice is the selection of the mutation probabilities, as the most appropriate mutation rate depends on the fitness of the starting model. Another consideration is that in general, larger models require lower mutation probabilities to allow fine-tuning of the well-performing models. However, any mutation probability in the range of 0.01-0.1 can sufficiently refine the model and reach saturation with enough iterations. We found that the other parameters have negligible impact on the performance. When an equal number of models were generated, the number of iterations did not make a significant difference as long as it was over a certain threshold, i.e., larger than ten when 100 models are generated. Similarly, the number of models kept to the next iteration did not make a significant difference as long as it was comparable to the number of models generated for each iteration, i.e., lower than five when five models are generated at each iteration. The optimal number of models to generate using mix (crossing) is small, i.e., one for the sampled models when five models were generated for each iteration.
Methods of the benchmark analysis to test the overall performance of boolmore
Following the common practice in such benchmarks, we used existing Boolean models as ground truth. We used the original model’s interaction graph to generate a randomized starting model to be refined. We used the minimal trap spaces of the original model under various perturbations to generate the artificial perturbation-observation pairs (which we will refer to as artificial experiments). We used 80% of the artificial experiments as the training set to refine a randomized starting model and used the remaining 20% as the validation set to test the accuracy of the refined model in recapitulating newly encountered experimental results.
We used all Boolean models with 30 or fewer nodes available in the Cell Collective (a repository of peer-reviewed, experimentally supported Boolean models37), leading to 40 models. For each model, we ran five replicate benchmark runs using a unique starting model and a unique set of artificial experiments.
We generated five starting models for each model by randomizing the binary representation of the Boolean functions of the original model. This randomization keeps the functions monotonic and consistent with the original interaction graph, but may yield fewer regulators than the original. The missing regulators can be added back in during the iterations of the model refinement process.
For each model with N nodes, we generated five sets of 10*N artificial experiments, 80% of which were used for training, for a coverage that is comparable to that of our case study (505 experiments for 68 not fully constrained nodes). Each artificial experiment consists of a set of nodes whose state is controlled (kept fixed) and a node whose state is observed. We aimed to select the controlled and observed nodes such that the collection of artificial experiments is representative of empirical perturbation-observation pairs.
The controlled set of nodes always included the source nodes of the network, which describe the signals and experimental context. Additional non-source nodes were included in the controlled sets such that their number followed a decreasing frequency (such that the majority of control sets included a single non-source node). This decreasing frequency reflects the lower representation of combinatorial perturbations due to the difficulty of their practical implementation.
We ensured that the sink nodes, which represent phenotypes in most models, are observed for each unique controlled set of nodes. We also assigned more observations to the smaller controlled sets (such as the wild type), reflecting the real-world dataset of our case study. We fixed the values of the nodes in the controlled set randomly and determined the average node value of the observed node in the minimal trap spaces. Depending on the average value, the result was classified into one of the five categories described previously, using thresholds that ensure that the original model would have a perfect fitness score.
We used boolmore to refine the models over 100 iterations and score them by comparing their results to the results of a subset (80%) of the artificial experiments following the procedure described earlier.
Data availability
The datasets generated and analyzed during the current study are available in the github repository https://github.com/kyuhyongpark/boolmore.
Code availability
The Python package boolmore and the training/validation datasets for this study are available in the github repository https://github.com/kyuhyongpark/boolmore.
References
Abou-Jaoudé, W. et al. Logical modeling and dynamical analysis of cellular networks. Front. Genet. 7, 94 (2016).
McCulloch, W. S. & Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biophys. 5, 115–133 (1943).
Hopfield, J. J. Neural networks and physical systems with emergent collective computational abilities. Proc. Natl. Acad. Sci.79, 2554–2558 (1982).
Castellano, C., Fortunato, S. & Loreto, V. Statistical physics of social dynamics. Rev. Mod. Phys. 81, 591–646 (2009).
Campbell, C., Yang, S., Albert, R. & Shea, K. A network model for plant-pollinator community assembly. Proc. Natl. Acad. Sci.108, 197–202 (2011).
Veitia, R. A. A sigmoidal transcriptional response: cooperativity, synergy and dosage effects. Biol. Rev. Camb. Philos. Soc. 78, 149–170 (2003).
Tyson, J. J., Chen, K. C. & Novak, B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 15, 221–231 (2003).
Martinez-Sanchez, M. E., Mendoza, L., Villarreal, C. & Alvarez-Buylla, E. R. A minimal regulatory network of extrinsic and intrinsic factors recovers observed patterns of CD4+T cell differentiation and plasticity. PLoS Comput. Biol. 11, e1004324 (2015).
Steinway, S. N. et al. Combinatorial interventions inhibit TGFβ-driven epithelial-to-mesenchymal transition and support hybrid cellular phenotypes. npj Syst. Biol. Appl 1, 15014 (2015).
Rodríguez-Jorge, O. et al. Cooperation between T cell receptor and Toll-like receptor 5 signaling for CD4+T cell activation. Sci. Signal. 12, eaar3641 (2019).
Gómez Tejeda Zañudo, J. et al. Cell line–specific network models of ER+ breast cancer identify potential PI3Kα inhibitor resistance mechanisms and drug combinations. Cancer Res. 81, 4603–4617 (2021).
Rozum, J. C., Deritei, D., Park, K. H., Zañudo, J. G. T. & Albert, R. Pystablemotifs: Python library for attractor identification and control in Boolean networks. Bioinformatics 38, 1465–1466 (2021).
Klarner, H., Streck, A. & Siebert, H. PyBoolNet: a python package for the generation, analysis and visualization of boolean networks. Bioinformatics 33, 770–772 (2017).
Beneš, N. et al. AEON.py: Python library for attractor analysis in asynchronous Boolean networks. Bioinformatics 38, 4978–4980 (2022).
Paulevé, L., Kolčák, J., Chatain, T. & Haar, S. Reconciling qualitative, abstract, and scalable modeling of biological networks. Nat. Commun. 11, 4256 (2020).
Naldi, A. et al. The CoLoMoTo interactive notebook: accessible and reproducible computational analyses for qualitative biological networks. Front. Physiol. 9, 680 (2018).
Trinh, V.-G., Benhamou, B. & Soliman, S. Trap spaces of Boolean networks are conflict-free siphons of their Petri net encoding. Theor. Comput. Sci. 971, 114073 (2023).
Trinh, H.-C. & Kwon, Y.-K. A novel constrained genetic algorithm-based Boolean network inference method from steady-state gene expression data. Bioinformatics 37, i383–i391 (2021).
Dorier, J. et al. Boolean regulatory network reconstruction using literature based knowledge with a genetic algorithm optimization method. BMC Bioinforma. 17, 410 (2016).
Ghaffarizadeh, A., Podgorski, G. J. & Flann, N. S. Applying attractor dynamics to infer gene regulatory interactions involved in cellular differentiation. Biosystems 155, 29–41 (2017).
Müssel, C. et al. CANTATA-prediction of missing links in Boolean networks using genetic programming. Bioinformatics 38, 4893–4900 (2022).
Palli, R., Palshikar, M. G. & Thakar, J. Executable pathway analysis using ensemble discrete-state modeling for large-scale data. PLoS Comput. Biol. 15, e1007317 (2019).
Muñoz, S., Carrillo, M., Azpeitia, E. & Rosenblueth, D. A. Griffin: a tool for symbolic inference of synchronous boolean molecular networks. Front. Genet. 9, 39 (2018).
Woodhouse, S., Piterman, N., Wintersteiger, C. M., Göttgens, B. & Fisher, J. SCNS: a graphical tool for reconstructing executable regulatory networks from single-cell genomic data. BMC Syst. Biol. 12, 59 (2018).
Chevalier, S., Froidevaux, C., Paulevé, L. & Zinovyev, A. Synthesis of Boolean Networks from Biological Dynamical Constraints using Answer-Set Programming. in Proc. IEEE 31st International Conference on Tools with Artificial Intelligence 34–41 (IEEE, 2019).
Stoeger, T., Gerlach, M., Morimoto, R. I. & Nunes Amaral, L. A. Large-scale investigation of the reasons why potentially important genes are ignored. PLoS Biol. 16, e2006643 (2018).
Begley, C. G. & Ellis, L. M. Drug development: raise standards for preclinical cancer research. Nature 483, 531–533 (2012).
Li, S., Assmann, S. M. & Albert, R. Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biol. 4, e312 (2006).
Albert, R. et al. A new discrete dynamic model of ABA-induced stomatal closure predicts key feedback loops. PLoS Biol. 15, e2003451 (2017).
Maheshwari, P., Du, H., Sheen, J., Assmann, S. M. & Albert, R. Model-driven discovery of calcium-related protein-phosphatase inhibition in plant guard cell signaling. PLoS Comput. Biol. 15, e1007429 (2019).
Maheshwari, P., Assmann, S. M. & Albert, R. A guard cell abscisic acid (ABA) network model that captures the stomatal resting state. Front. Physiol. 11, 927 (2020).
Mitchell, M. An Introduction to Genetic Algorithms (MIT Press, London)
Terfve, C. et al. CellNOptR: a flexible toolkit to train protein signaling networks to data using multiple logic formalisms. BMC Syst. Biol. 6, 133 (2012).
Flobak, Å et al. Fine tuning a logical model of cancer cells to predict drug synergies: combining manual curation and automated parameterization. Front. Syst. Biol. 3, 1252961 (2023).
Gouveia, F., Lynce, I. & Monteiro, P. T. Revision of boolean models of regulatory networks using stable state observations. J. Comput. Biol. 27, 144–155 (2020).
Aleixo, F., Knorr, M. & Leite, J. Revising Boolean logical models of biological regulatory networks. in Proc. 20th International Conference on Principles of Knowledge Representation and Reasoning 19 12–22 (IJCAI, 2023).
Helikar, T. et al. The cell collective: toward an open and collaborative approach to systems biology. BMC Syst. Biol. 6, 96 (2012).
Waidyarathne, P. & Samarasinghe, S. Boolean calcium signalling model predicts calcium role in acceleration and stability of abscisic acid-mediated stomatal closure. Sci. Rep. 8, 17635 (2018).
Maheshwari, P., Assmann, S. M. & Albert, R. Inference of a boolean network from causal logic implications. Front. Genet. 13, 836856 (2022).
Eisinger, W., Ehrhardt, D. & Briggs, W. Microtubules are essential for guard-cell function in Vicia and Arabidopsis. Mol. Plant 5, 601–610 (2012).
Yang, H.-M., Zhang, J.-H. & Zhang, X.-Y. Regulation mechanisms of stomatal oscillation. J. Integr. Plant Biol. 47, 1159–1172 (2005).
Everitt, B. S. & Skrondal, A. The Cambridge Dictionary of Statistics. (Cambridge Univ. Press, 2010).
Van Ham, P. How to deal with variables with more than two levels. in Kinetic Logic A Boolean Approach to the Analysis of Complex Regulatory Systems 326–343 (Springer, Heidelberg, 1979).
Didier, G., Remy, E. & Chaouiya, C. Mapping multivalued onto Boolean dynamics. J. Theor. Biol. 270, 177–184 (2011).
Mony, V. K. et al. Context-specific regulation of lysosomal lipolysis through network-level diverting of transcription factor interactions. Proc. Natl. Acad. Sci.118, e2104832118 (2021).
Emmert-Streib, F., Dehmer, M. & Haibe-Kains, B. Gene regulatory networks and their applications: understanding biological and medical problems in terms of networks. Front Cell Dev. Biol. 2, 38 (2014).
Nguyen, H., Tran, D., Tran, B., Pehlivan, B. & Nguyen, T. A comprehensive survey of regulatory network inference methods using single cell RNA sequencing data. Brief. Bioinform. 22, bbaa190 (2021).
Pillich, R. T. et al. NDEx: accessing network models and streamlining network biology workflows. Curr. Protoc. 1, e258 (2021).
Licata, L. et al. SIGNOR 2.0, the signaling network open resource 2.0: 2019 update. Nucleic Acids Res. 48, D504–D510 (2020).
Milacic, M., et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 52, 672–678 (2023).
Klamt, S., Saez-Rodriguez, J., Lindquist, J. A., Simeoni, L. & Gilles, E. D. A methodology for the structural and functional analysis of signaling and regulatory networks. BMC Bioinform. 7, 56 (2006).
Park, K. H., Costa, F. X., Rocha, L. M., Albert, R. & Rozum, J. C. Models of cell processes are far from the edge of chaos. PRX Life 1, 023009 (2023).
Shmulevich, I. & Dougherty, E. R. Probabilistic Boolean Networks. (Soc. Ind. Appl. Math., 2010).
Murrugarra, D., Veliz-Cuba, A., Aguilar, B., Arat, S. & Laubenbacher, R. Modeling stochasticity and variability in gene regulatory networks. EURASIP J. Bioinform. Syst. Biol. 2012, 5 (2012).
Holtzapple, E., Telmer, C. A. & Miskov-Zivanov, N. FLUTE: fast and reliable knowledge retrieval from biomedical literature. Database 2020, baaa056 (2020).
Ahmed, Y., Telmer, C. A. & Miskov-Zivanov, N. CLARINET: efficient learning of dynamic network models from literature. Bioinforma. Adv. 1, vbab006 (2021).
Kadelka, C. et al. A meta-analysis of Boolean network models reveals design principles of gene regulatory networks. Sci. Adv. 10, eadj0822 (2024).
Azpeitia, E., Weinstein, N., Benítez, M., Mendoza, L. & Alvarez-Buylla, E. R. Finding missing interactions of the arabidopsis thaliana root stem cell niche gene regulatory network. Front. Plant Sci. 4, 110 (2013).
Acknowledgements
We are grateful for the advice of Prof. István Albert, Prof. Sarah M. Assmann, Prof. Dezhe Jin, and the helpful suggestions of Dr. Xiao Gan, Dr. Fatemeh Sadat Fatemi Nasrollahi, and Dr. Eli Newby. This study was funded by the National Science Foundation under grants MCB 1715826 and IIS 1814405. The funder played no role in study design, data collection, analysis, and interpretation of data, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
K.H.P. developed and implemented the algorithms, performed the benchmark analyses and case study, and was a major contributor in writing the manuscript. J.C.R. developed the binary representation of the functions used for mutations and advised the project. R.A. assisted with the biological aspects of the case study, advised the project, and was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
K.H.P. and J.C.R. declare no financial or non-financial competing interests. R.A. serves as Associate Editor of this journal and has no role in the peer-review or decision to publish this manuscript. R.A. declares no financial competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Park, K.H., Rozum, J.C. & Albert, R. Automated model refinement using perturbation-observation pairs. npj Syst Biol Appl 11, 65 (2025). https://doi.org/10.1038/s41540-025-00532-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41540-025-00532-y