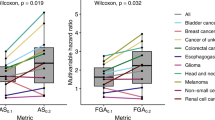
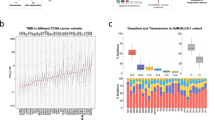
Abstract
Tumors with high copy number alteration (CNA) burden often respond poorly to immune checkpoint inhibitor therapy. However, how CNAs affect the anti-cancer immune response remains unclear. To address this, we set out to capture the transcriptional effects of CNAs and define a comprehensive landscape of immune-related transcriptional patterns. Hereto, we applied consensus independent component analysis to 294,159 bulk transcriptomic profiles. We demonstrated the predictive power of these patterns for immunotherapy response, their reproducibility across platforms, and their applicability to bulk, single-cell, and spatial transcriptomic data. Our analysis identified both novel inverse and positive associations between high CNA burden and immune-related transcriptional patterns across various cancer types. For example, higher CNA burden correlated with increased immunosuppression, including IL-17-producing cells and regulatory T cells. This resource, along with the classification of these transcriptional patterns as immune-suppressive and immune-stimulatory, may provide insights to improve immunotherapy efficacy in tumors with high CNA burden.
Similar content being viewed by others
Data availability
The CNA-immune associations can be explored at http://cna-immune.opendatainscience.net. All data, results, and codes generated as part of this study are available at https://zenodo.org/records/17715512. Gene expression data were collected from three public data repositories: Gene Expression Omnibus with accession number GPL570 generated with microarray Affymetrix HG-U133 Plus 2.0 (https://www.ncbi.nlm.nih.gov/geo/), ARCHS4 RNA-seq from human version v1.10 (https://maayanlab.cloud/archs4/download.html), and TCGA RNA-seq data from the Broad GDAC Firehose portal (https://gdac.broadinstitute.org/). ICI response datasets were obtained from the referenced studies. Single-cell RNA-seq data were obtained from the single-cell tumor immune atlas for precision oncology (https://zenodo.org/records/5205544). Spatial transcriptomic profiles were obtained from the 10x Genomics website (https://www.10xgenomics.com).
References
Chen, D. S. & Mellman, I. Elements of cancer immunity and the cancer–immune set point. Nature 541, 321–330 (2017).
Haslam, A. & Prasad, V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw. Open 2, e192535 (2019).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Liu, L. et al. Combination of TMB and CNA stratifies prognostic and predictive responses to immunotherapy across metastatic cancer. Clin. Cancer Res. 25, 7413–7423 (2019).
Chen, M. et al. Cold and heterogeneous T cell repertoire is associated with copy number aberrations and loss of immune genes in small-cell lung cancer. Nat. Commun. 12, 6655 (2021).
Lu, Z. et al. Tumor copy-number alterations predict response to immune-checkpoint-blockade in gastrointestinal cancer. J. Immunother. Cancer 8, e000374 (2020).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
Roh, W. et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 9, eaah3560 (2017).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
Safonov, A. et al. Immune gene expression Is associated with genomic aberrations in breast cancer. Cancer Res. 77, 3317–3324 (2017).
Balli, D., Rech, A. J., Stanger, B. Z. & Vonderheide, R. H. Immune cytolytic activity stratifies molecular subsets of human pancreatic cancer. Clin. Cancer Res. 23, 3129–3138 (2017).
Thapa, B. et al. The immune microenvironment, genome-wide copy number aberrations, and survival in mesothelioma. J. Thorac. Oncol. 12, 850–859 (2017).
Buccitelli, C. et al. Pan-cancer analysis distinguishes transcriptional changes of aneuploidy from proliferation. Genome Res. 27, 501–511 (2017).
Bhattacharya, A. et al. Transcriptional effects of copy number alterations in a large set of human cancers. Nat. Commun. 11, 715 (2020).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41, D991–D995 (2013).
Lachmann, A. et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 9, 1366 (2018).
Weinstein, J. N. et al. The Cancer Genome Atlas Pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Pusztai, L. et al. Durvalumab with olaparib and paclitaxel for high-risk HER2-negative stage II/III breast cancer: results from the adaptively randomized I-SPY2 trial. Cancer Cell 39, 989–998 (2021).
Wolf, D. M. et al. Redefining breast cancer subtypes to guide treatment prioritization and maximize response: predictive biomarkers across 10 cancer therapies. Cancer Cell 40, 609–623 (2022).
Cho, J.-W. et al. Genome-wide identification of differentially methylated promoters and enhancers associated with response to anti-PD-1 therapy in non-small cell lung cancer. Exp. Mol. Med. 52, 1550–1563 (2020).
Cho, J. W. et al. Genome-wide methylation patterns predict clinical benefit of immunotherapy in lung cancer. Clin. Epigenetics 12, 119 (2020).
Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949 (2017).
Cui, C. et al. Ratio of the interferon-γ signature to the immunosuppression signature predicts anti-PD-1 therapy response in melanoma. NPJ Genom. Med. 6, 7 (2021).
Gide, T. N. et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 35, 238–255 (2019).
Van Allen, E. M. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015).
Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019).
Kim, S. T. et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 24, 1449–1458 (2018).
Mariathasan, S. et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018).
Zhao, J. et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat. Med. 25, 462–469 (2019).
Braun, D. A. et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat. Med. 26, 909–918 (2020).
Borish, L. C. & Steinke, J. W. 2. Cytokines and chemokines. J. Allergy Clin. Immunol. 111, S460–S475 (2003).
Ricciotti, E. & FitzGerald, G. A. Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 31, 986–1000 (2011).
Ma, E. H. et al. Serine is an essential metabolite for effector T cell expansion. Cell Metabolism 25, 345–357 (2017).
Urzúa-Traslaviña, C. G. et al. Improving gene function predictions using independent transcriptional components. Nat. Commun. 12, 1464 (2021).
Nieto, P. et al. A single-cell tumor immune atlas for precision oncology. Genome Res. 31, 1913–1926 (2021).
Kandala, et al. Chronic chromosome instability induced by Plk1 results in immune suppression in breast cancer. Cell Rep. 42, 113266 (2023).
Li, J. et al. Non-cell-autonomous cancer progression from chromosomal instability. Nature 620, 1080–1088 (2023).
Wertheimer, T. et al. IL-23 stabilizes an effector Treg cell program in the tumor microenvironment. Nat. Immunol. 25, 512–524 (2024).
Castle, J. C. et al. Immunomic, genomic and transcriptomic characterization of CT26 colorectal carcinoma. BMC Genomics 15, 190 (2014).
Liu, C. et al. Blocking IL-17A enhances tumor response to anti-PD-1 immunotherapy in microsatellite stable colorectal cancer. J. Immunother. Cancer 9, e001895 (2021).
Kridin, K., Abdelghaffar, M., Mruwat, N., Ludwig, R. J. & Thaçi, D. Are interleukin 17 and interleukin 23 inhibitors associated with malignancies?—Insights from an international population-based study. J. Eur. Acad. Dermatol. Venereol. 38, 315–324 (2024).
de Vries, N. L. et al. γδ T cells are effectors of immunotherapy in cancers with HLA class I defects. Nature 613, 743–750 (2023).
Lerner, E. C. et al. CD8+ T cells maintain killing of MHC-I-negative tumor cells through the NKG2D–NKG2DL axis. Nat. Cancer 4, 1258–1272 (2023).
Beck, J. D. et al. Long-lasting mRNA-encoded interleukin-2 restores CD8+ T cell neoantigen immunity in MHC class I-deficient cancers. Cancer Cell 42, 568–582.e11 (2024).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Jassal, B. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 48, D498–D503 (2020).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Chiappetta, P., Roubaud, M. C. & Torrésani, B. Blind source separation and the analysis of microarray data. J. Comput. Biol. 11, 1090–1109 (2004).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Fernández-Delgado, M., Cernadas, E., Barro, S. & Amorim, D. Do we need hundreds of classifiers to solve real world classification problems?. J Mach. Learn. Res. 15, 3133–3181 (2014).
Webber, W., Moffat, A. & Zobel, J. A similarity measure for indefinite rankings. ACM Trans. Inf. Syst. 28, 1–38 (2010).
Acknowledgements
This work was supported by a grant from the Hanarth Fund to R.S.N.F., and by a KWF Kankerbestrijding grant [grant number KWF-14516] awarded to M.A.T.M.v.V. and R.S.N.F.
Author information
Authors and Affiliations
Contributions
R.S.N.F. conceived and designed this study. S.L., A.B., C.G.U.-T., and R.S.N.F. collected and assembled data and performed data analyses. S.L. and C.G.U.-T. developed the figures used in this study. S.L., A.B., C.G.U.-T., M.A.T.M.v.V., M.d.B., and R.S.N.F. contributed to the data interpretation, writing of the paper, and the final decision to submit the paper.
Corresponding author
Ethics declarations
Competing interests
M.d.B. received grants from the Dutch Cancer Society (KWF), the European Research Council (ERC), Health Holland (HH), Mendus, BioNovion, Aduro Biotech, Vicinivax, Genmab, and IMMIOS (all paid to the institute); received non-financial support from BioNTech, Surflay Nanotec, and Merck Sharp & Dohme; is a stock option holder in Sairopa; all unrelated to the submitted work. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Loipfinger, S., Bhattacharya, A., Urzúa-Traslaviña, C.G. et al. Association of copy number alterations with the immune transcriptomic landscape in cancer. npj Syst Biol Appl (2026). https://doi.org/10.1038/s41540-026-00649-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41540-026-00649-8