Abstract
The COVID-19 pandemic spurred mRNA vaccine innovation, but new SARS-CoV-2 variants highlight the need for vaccines with improved potency and durability. This report presents a novel mRNA vaccine platform encoding virus-like particle antigens (mRNA-VLPs) that mimic native virus structures, aiming to boost antibody responses via enhanced B cell activation. In animal studies, mRNA-VLP vaccines generated stronger neutralizing antibody responses across multiple variants compared to conventional mRNA vaccines expressing native spike proteins. In non-human primates, these elevated antibodies lasted at least six months. An mRNA-VLP vaccine encoding the Omicron spike outperformed traditional mRNA vaccines in mice as both a monovalent and bivalent (with ancestral spike) formulation. In hamsters, even low doses of mRNA-VLP vaccine provided complete protection, similar to high doses of native spike mRNA vaccines. These results suggest the mRNA-VLP platform could significantly strengthen vaccine efficacy and breadth against evolving SARS-CoV-2 variants.
Similar content being viewed by others
Introduction
The widespread morbidity and mortality associated with the coronavirus disease 2019 (COVID-19) pandemic precipitated the most extensive and rapid global vaccine development program in history against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus. SARS-CoV-2 variants with distinct phenotypic characteristics, including increased transmissibility and severity along with the development of immune evasion, began to emerge approximately 8 months into the pandemic1. Thus, along with declining antibody levels2, the durability of protection waned3. As SARS-CoV-2 continues to evolve, there is an ongoing need for rapid development and evaluation of new, more broadly active SARS-CoV-2 vaccines with improved durability of protection.
The COVID-19 pandemic also saw the first mRNA vaccine achieve full FDA approval in the US4. mRNA vaccines typically comprise mRNA strands encapsulated in a lipid nanoparticle (LNP)5. Following internalization of the LNP into cells and mRNA release, the mature viral protein or related antigens are expressed on the host cell surface, where they can elicit a protective immune response. The antigens can also be secreted to induce an immune response by activating antigen-presenting cells; this mechanism has the advantage of activating both the innate and adaptive immune systems, thereby generating both humoral and cellular responses6. mRNA vaccines have several advantages over other types of vaccines including: (1) their development is relatively fast as mRNA can be made within weeks of identifying and sequencing the pathogen and corresponding target antigen gene7, (2) mRNAs encoding different antigens are chemically and physically similar and the same formulation, design, and manufacturing process can be applied to new/updated sequences5, and (3) revaccination is not limited by an anti-vector response.
A vaccine platform combining an LNP with mRNA that expresses virus-like particle antigens (mRNA-VLP) is being developed to increase the potency, breadth, and durability of the neutralizing antibody (nAb) response against SARS-CoV-2, building upon similar approaches to deliver vaccine antigens as VLPs via mRNA8 or plasmid DNA9. mRNA-based VLPs are multiprotein structures that can assemble into particles to display multiple antigens mimicking the organization and conformation of native viruses but lacking the viral genome. The multimeric antigen display on VLPs enables the potential for more potent stimulation of the immune system through cross-linking of B cell receptors10. Some VLPs also have the capacity to present vaccine antigens from multiple viral variants to the immune system11. Furthermore, protein-based VLPs have already been used in the development of several successful vaccines12,13.
The aim of this report is to demonstrate that the mRNA-VLP vaccine platform shows increased potency, breadth, and durability of the SARS-CoV-2 nAb response in animal models compared to the same spike antigen expressed as a cell surface, membrane-anchored protein like those in licensed SARS-CoV-2 mRNA vaccines. Toward this end, we constructed the mRNA-VLP Delta vaccine that contains an mRNA encoding the Delta variant of the SARS-CoV-2 spike glycoprotein fused to the Helicobacter pylori (H. pylori) ferritin scaffold protein sequence. The self-assembling H. pylori ferritin scaffold has been used successfully to present many different viral antigens14, including the influenza hemagglutinin protein, both preclinically15 and in human clinical trials16,17. We show that mRNA-VLP Delta vaccine elicits enhanced virus nAb responses across multiple SARS-CoV-2 variants in both mice and non-human primates (NHP) when compared to mRNA expressing the full-length Delta spike as a native membrane-anchored protein (referred to as mRNA-native Delta). In addition, we demonstrate that the higher nAb responses induced by mRNA-VLP Delta were maintained for at least 6 months in NHP. Furthermore, we extend the results by showing that an mRNA-VLP vaccine encoding the Omicron BA.4/5 spike elicits higher nAb responses in mice either as a monovalent vaccine or when administered in combination with an mRNA-VLP encoding the ancestral D614G spike (bivalent mRNA-VLP composition) compared to a bivalent vaccine expressing the same variant spike sequences as native spike proteins. Lastly, the vaccine efficacy of the mRNA-VLP platform was confirmed in a hamster model of SARS-CoV-2 virus challenge, wherein low doses of the mRNA-VLP vaccine afforded complete protection, comparable to that of a high dose of the mRNA vaccine expressing native spike protein antigens.
Results
Development and evaluation of the mRNA-VLP vaccine using a prototypical Delta spike antigen
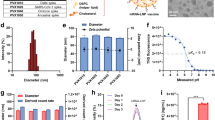
To determine whether spike-ferritin mRNA expressed in HEP2 cells formed VLPs following transfection, we employed cryo-EM to determine the 3-dimensional structure of the released and purified spike-ferritin proteins. Cryo-EM structures revealed a high-resolution (2.2 Å) ferritin core with eight Delta spike trimers at the threefold axes of the ferritin cage (Fig. 1A, B). Whilst the core showed clear features consistent with this resolution, density corresponding to the spike at the threefold axes showed that the spike is highly flexible on the surface of the cage. Using the symmetry of the cage and focused refinements around the low-resolution spike density, three high-resolution structures were determined, ranging from 3.1 to 3.3 Å in resolution. The resolved spike structures confirmed that the spike can fold and is glycosylated in a native manner on the ferritin nanoparticle in a way that is identical to the spike found on the viral surface (Fig. 1)18. Three distinct conformations of the spike protein were observed (Fig. 1C). The first was in the closed conformation with all receptor binding domains (RBD) in the down position and the trimer adopting a canonical threefold symmetry. The additional 2 conformations showed varying flexibility in the RBDs with one structure displaying 1 RBD in the “up” conformation and the other with 2 RBDs in the “up” conformation.
A Composite map of the Delta spike-ferritin VLP was constructed through docking the high-resolution ferritin (map (gray) and spike maps (gold) into the low-resolution consensus map. B Close-up view of one trimer on the VLP showing the assembly is comprised of 8 trimers of spike-ferritin fusion protomers (colored by protomer). C Models corresponding to the three conformations of spike observed on individual RBDs, showing all RBDs closed (left), one RBD in the up position and two RBDs down (center), and two RBDs up and one RBD down (right) conformations. Each protomer is individually colored with glycans highlighted in cyan. SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, RBD receptor binding domain of the spike protein, VLP virus-like particle.
To compare the magnitude and breadth of nAb levels elicited by an mRNA-VLP versus an mRNA-native spike antigen, SARS-CoV-2 naive female BALB/c mice were injected with phosphate-buffered saline (PBS), mRNA-native Delta, or mRNA-VLP Delta on day 1 and day 28 and sera collected on day 42 to test against a panel of SARS-CoV-2 pseudoviruses (Fig. 2A). The breadth of nAbs was specifically tested against a diverse panel including ancestral (Delta and Wuhan_D614G) and early Omicron (BA.1, BA.2, BA.4/5, and in later experiments BQ.1.1) variants (Fig. 2B). Immunization with mRNA-native Delta vaccine generated nAb levels against D614G and Delta pseudoviruses with geometric mean titers (GMTs) of 3095 and 5789, respectively (Fig. 2C); lower levels of nAbs were elicited against the BA.1, BA.2, or BA.4/5 pseudoviruses with GMTs ranging from 112 to 232. Mice immunized with mRNA-VLP Delta vaccine elicited elevated nAb levels against all pseudoviruses tested with higher GMTs in comparison to the mRNA-native Delta responses: D614G: 5-fold; Delta: 3-fold; BA.1: 49-fold; BA.2: 66-fold; and BA.4/5: 24-fold (Fig. 2C).
A Groups of naive BALB/c mice (n = 4–6 /group) were administered 1.0 μg of LNP-formulated mRNA vaccine (mRNA-native Delta or mRNA-VLP Delta) or PBS control intramuscularly on study days 1 and 28. Fourteen days following the second dose (study day 42), sera were collected and pseudovirus-based neutralization assays performed using a panel of SARS-CoV-2 pseudoviruses (pV) as indicated. B Phylogenetic tree of SARS-CoV-2 spike sequences. C Day 42 serum anti-SARS-CoV-2 neutralization antibody titers. The geometric mean titer (GMT) ± geometric standard deviation is shown with indicated p value comparing the mRNA-native and mRNA-VLP responses for each tested pV by the Wilcoxon–Mann–Whitney test; the dotted horizontal line indicates the assay lower limit of detection (LLD). Ab antibody, D614G ancestral D614G spike, ID50 half-maximal inhibitory dilution, LNP lipid nanoparticle, native full-length Delta spike protein containing the S2P pre-fusion spike stabilization mutations, ns not significant, PBS phosphate-buffered saline, pV pseudovirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, VLP virus-like particle.
To determine whether the improved immunogenicity of the VLP-based vaccine observed in BALB/c mice also occurred in NHP, the two different vaccines were tested in SARS-CoV-2 naive cynomolgus macaques at a dose of 10 µg (Fig. 3A). The mRNA-VLP Delta vaccine elicited nAb levels that were significantly higher (p < 0.01) against all pseudoviruses when compared with the mRNA-native Delta vaccine two weeks post second immunization (D614G: 15-fold; Delta: 9-fold; BA.1: 29-fold; BA.2: 18-fold; BA.4/5: 8-fold; Fig. 3B). Although not statistically significant, the mRNA-VLP Delta vaccine exhibited a trend toward increased numbers of spike-specific B cells in the periphery of vaccinated animals (Fig. 3C and Supp Fig. 1). Intracellular cytokine staining (ICS) of peripheral blood CD4+ T cells following spike peptide stimulation revealed an exclusive Th1 CD4+ T cell response for both vaccines with undetectable levels of Th2 CD4+ T cells (Fig. 3D and Supp Fig. 1). Similar to previous reports describing immunogenicity of COVID mRNA vaccines in NHPs19, no detectable levels of spike-specific CD8+ T cells were measured following ICS/flow cytometry of spike peptide stimulated cells (data not shown).
A Two groups of NHP (n = 6/group) were administered 10 μg of LNP-formulated mRNA vaccine (mRNA-native Delta or mRNA-VLP Delta) intramuscularly on study days 1 and 28. B Fourteen days following the second dose (study day 42), sera were collected and pseudovirus-based neutralization assays performed using a panel of SARS-CoV-2 pseudoviruses as indicated. Day 42 serum anti-SARS-CoV-2 neutralization antibody titers. The geometric mean titer (GMT) ± geometric standard deviation is shown with indicated p value comparing the mRNA-native and mRNA-VLP responses for each tested pseudovirus (pV) by the Wilcoxon-Mann-Whitney test; the dotted horizontal line indicates the assay lower limit of detection (LLD). C On study days 27 and 56, spike-specific IgG+ B cell frequencies were measured in PBMCs using flow cytometry following staining with a panel of antibodies specific for B cell markers and a fluorescent spike probe. D On study days 27 and 56, spike-specific memory CD4 + T cell frequencies and phenotypes were measured in the periphery using flow cytometry following ex vivo stimulation of PBMCs with a spike-specific overlapping peptide pool and intracellular cytokine staining for Th1 (IFNγ, IL-2, TNFα) and Th2 (IL-4, IL-13) cytokines. In the box-whisker plots, horizontal bars indicate group median, boxes are the interquartile range, and whiskers are the range. E Sera were collected at different timepoints post vaccination (as indicated) and pV-based neutralization assays were performed to evaluate anti-SARS-CoV-2 (Delta variant) neutralizing antibody titers shown as the GMT for each group with the indicated p value for each timepoint. F Approximately 196 days after the first immunization, bone marrow aspirates were processed to enumerate long-lived antibody-secreting cells among individual animals by ELISpot assay. Data are represented as the number of spot-forming cells (SFCs) per 1 × 106 bone marrow cells plated with determined p value indicated. Ab antibody, D614G ancestral D614G spike, ID50 half-maximal inhibitory dilution, native Delta full-length Delta spike protein containing the S2P pre-fusion spike stabilization mutations, ns not significant, PBMCs peripheral blood mononuclear cells.
We also wanted to determine whether the nAb levels elicited by mRNA-VLP vaccine were more durable than those elicited by the mRNA-native vaccine. Serum samples were tested for nAb titers out to day 196 (Fig. 3E). These data revealed that the higher nAb levels elicited by the mRNA-VLP Delta vaccine remained elevated for at least 5.5 months after the primary vaccine series in comparison to the mRNA-native Delta vaccine (Fig. 3E). The mRNA-VLP Delta and mRNA-native Delta groups had nAb GMTs of 236 and 68 (p = 0.0238), respectively, at 196 days following the first vaccine administration (Fig. 3E). Lastly, to determine whether these increased nAb levels at 6.5 months corresponded with memory B cell responses, spike-specific antibody secreting cells in the bone marrow (BM) 168 days after the second immunization were measured (Fig. 3F). The mRNA-VLP Delta and mRNA-native Delta vaccines induced a mean of 16.94 and 8.82 (p = 0.038), respectively, of spike-specific long-lived antibody secreting cells per 106 BM cells.
Evaluation of mRNA-VLP vaccine against Omicron BA.4/5 variants
Next, we evaluated whether the mRNA-VLP vaccine was also immunogenic against the Omicron BA.4/5 variant that was included in the 2023–2024 winter vaccine update. We developed mRNA-VLP constructs with both the ancestral Wuhan_D614G spike and the Omicron BA.4/5 spike for immunogenicity testing in mice. The two mRNA-VLP constructs were first administered separately and tested against a comparator mRNA-native construct encoding the native Omicron BA.4/5 spike protein in naive mice.
Administration of the mRNA-VLP D614G vaccine to naive mice elicited a robust nAb response against D614G and Delta with lower levels of nAbs against the Omicron BA.1, BA.2, BA.4/5, or BQ.1.1 pseudoviruses (Fig. 4). Immunization with mRNA-VLP BA.4/5 vaccine induced nAbs against BA.4/5 and the antigenically distinct BQ.1.1 that were similar (i.e., within twofold) to that of the mRNA-native BA.4/5 vaccine (Fig. 4). However, the mRNA-VLP BA.4/5 vaccine elicited a more potent nAb response against the remaining variants with 7, 4, 70, and 7-fold increases in GMTs against the D614G, Delta, BA.1, and BA.2 pseudoviruses, respectively, compared to the mRNA-native BA.4/5 vaccine (Fig. 4) demonstrating the expanded potency and breadth of the mRNA-VLP platform versus the mRNA-native vaccine.
Groups of naive BALB/c mice (n = 6/group) were administered 1.0 μg of LNP-formulated mRNA vaccine (mRNA-native BA.4/5, mRNA-VLP D614G, or mRNA-VLP BA.4/5) or PBS control intramuscularly on study days 1 and 21. Fourteen days following the second dose (study day 35), sera were collected and pseudovirus-based neutralization assays performed using a panel of SARS-CoV-2 pseudoviruses (pV) as indicated. The geometric mean titer (GMT) ± geometric standard deviation is shown with indicated p value comparing the different vaccine group responses for each tested pV by Kruskal–Wallis test, the dotted horizontal line indicates the assay lower limit of detection (LLD). Ab antibody, D614G ancestral D614G spike, ID50 half-maximal inhibitory dilution, native full-length spike protein containing the S2P pre-fusion spike stabilization mutations, ns not significant, PBS phosphate-buffered saline, pV pseudovirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, VLP virus-like particle; *p < 0.05; **p < 0.01; ***p < 0.001.
Since most individuals currently receiving SARS-CoV-2 vaccines have been previously vaccinated and/or infected, the mRNA vaccines were administered to groups of vaccine-primed mice to evaluate boosting capacity of virus nAb titers in a more relevant context (Fig. 5A). Approximately 144 days after primary vaccine series (two intramuscularly [IM] immunizations with mRNA-native D164G), mice were immunized with a single low dose amount (0.5 µg) of either bivalent mRNA-native vaccine or bivalent mRNA-VLP vaccine with each vaccine comprised of the ancestral Wuhan and Omicron BA.4/5 spike antigen sequences. The resulting nAb levels were measured two weeks later against a panel of variant pseudoviruses (Fig. 5B). The bivalent mRNA-native vaccine elicited nAb levels against the D614G, BA.1, and BA.2 pseudoviruses with GMTs of 16408, 2401, and 419, respectively, with low but measurable nAb titers against BA.4/5 and BQ.1.1 in some animals (Fig. 5B). Interestingly, higher nAb levels were elicited against both the antigenically matched BA.4/5 (23-fold higher) and the antigenically distinct BA.2 (5-fold higher) and BQ.1.1 (3-fold higher) viruses with the bivalent mRNA-VLP vaccine versus the bivalent mRNA-native vaccine (Fig. 5B). Although not statistically significant, these data suggest that the bivalent mRNA-VLP vaccine has increased potency against the antigenically diverse Omicron variants BA.2 and BQ.1.1 pseudoviruses when compared to a bivalent mRNA-native vaccine in previously immunized mice.
A Groups of naive BALB/c mice (n = 6/group) were administered two intramuscular immunizations (28 days apart) of 0.2 μg of an LNP-formulated mRNA vaccine encoding the ancestral D614G native spike protein and then allowed to rest for 144 days. After the rest period, the mice were administered a third intramuscular dose of LNP-formulated mRNA vaccine (0.5 μg): bivalent mRNA-native vaccine encoding the ancestral D614G and Omicron BA.4/5 spike proteins or a bivalent mRNA-VLP vaccine encoding the ancestral D614G and Omicron BA.4/5 spike proteins as VLP antigens. Fourteen days later, sera were collected and pseudovirus-based neutralization assays performed using a panel of SARS-CoV-2 pseudoviruses as indicated. B The geometric mean titer (GMT) ± geometric standard deviation is shown with indicated p value comparing the different vaccine group responses for each tested pseudovirus (pV) by Kruskal-Wallis test; the dotted horizontal line indicates the assay lower limit of detection (LLD). Ab antibody, D614G ancestral D614G spike, ID50 half-maximal inhibitory dilution, native spike full-length spike protein containing the S2P pre-fusion spike stabilization mutations, ns not significant, pV pseudovirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, VLP virus-like particle.
mRNA-VLP vaccine protects hamsters following a SARS-CoV-2 infection
Efficacy of the bivalent mRNA-VLP vaccine was tested in the well-characterized Syrian hamster model of SARS-CoV-2 infection20 with the ancestral Wuhan virus strain. Hamsters were immunized twice with control PBS, 10 μg of a bivalent mRNA-native vaccine encoding Wuhan_D614G and Omicron BA.4/5 spike antigens, and either 2 or 10 μg of the bivalent mRNA-VLP vaccine encoding Wuhan_D614G and Omicron BA.4/5 spike antigens as VLPs. Two weeks after the second immunization (day 35) and prior to intranasal challenge with the ancestral Wuhan virus strain (day 42), nAb levels against vaccine-matched pseudoviruses were measured on day 35 (Fig. 6A). The 10 μg dose of bivalent mRNA-native vaccine, 10 μg dose of bivalent mRNA-VLP vaccine, and 2 μg dose of bivalent mRNA-VLP vaccine all elicited comparable nAb levels to the ancestral Wuhan_D614G pseudovirus with GMTs of 2947, 2592, and 1581, respectively (Fig. 6B). However, both the 10 µg and 2 µg doses of bivalent mRNA-VLP vaccine generated higher and statistically significant (p < 0.001) levels of nAb to the Omicron BA.4/5 pseudovirus in comparison to the 10 μg of bivalent mRNA-native vaccine with nAb titers of 18648, 5702, and 373, respectively. The GMTs for the bivalent mRNA-VLP vaccine 10 μg and 2 μg groups were ~50- and 15-fold higher, respectively, than the nAb response elicited by 10 μg of the bivalent mRNA-native vaccine (Fig. 6B). These data show that the bivalent mRNA-VLP vaccine exhibits comparable potency at a 2 µg dose compared to a 10 µg dose of the bivalent mRNA-native vaccine in naive hamsters.
A Four groups of Syrian golden hamster (n = 16/group) were administered two intramuscular immunizations (21 days apart) of PBS control or LNP-formulated mRNA vaccines including 10 μg of a bivalent mRNA-native vaccine encoding the ancestral D614G and Omicron BA.4/5 spike proteins, and either 10 or 2 μg of the bivalent mRNA-VLP vaccine encoding the D614G spike VLP and Omicron BA.4/5 spike VLP. Fourteen days following the second immunization, sera were collected for pseudovirus-based neutralization assays. On study day 42, hamsters were challenged intranasally with ancestral SARS-CoV-2 virus and then weighed and monitored daily. One cohort of hamsters from each group (n = 8) was euthanized on study day 45 (three days post virus challenge), which correlates with peak virological measurements. A second cohort of hamsters from each group (n = 8) was euthanized on study day 49 (seven days post virus challenge), which correlates with peak pathological findings associated with SARS-CoV-2 infection. B The geometric mean neutralizing antibody titer (GMT) ± geometric standard deviation is shown for both ancestral D614G and Omicron BA.4/5 pseudoviruses; dotted horizontal line indicates the assay lower limit of detection (LLD). C Mean percent change in body weights (relative to weights recorded on day 42 prior to challenge) and standard deviation post virus challenge. D Oral swabs collected on day 2 post virus challenge were assessed for viral sgRNA concentrations by qRT-PCR. E Lung homogenates collected on days 3 and 7 post virus challenge were assessed for infectious virus by TCID50 assay. F Lung homogenates collected on days 3 and 7 post virus challenge were assessed for viral sgRNA concentrations by qRT-PCR. Data represent geometric means ± geometric standard deviations; dotted horizontal line indicates the assay LLD. p values comparing the different vaccine group responses are indicated as determined by Kruskal–Wallis test. ID50 half-maximal inhibitory dilution, ns not significant, PBS phosphate-buffered saline, qRT-PCR quantitative real-time polymerase chain reaction, sgRNA subgenomic RNA, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To examine whether the 10 µg and 2 µg doses of the bivalent mRNA-VLP vaccine elicited a protective immune response comparable to the bivalent mRNA-native vaccine, hamsters were challenged intranasally with SARS-CoV-2 on day 42 and body weight monitored daily until the scheduled necropsies were performed on study days 45 and 49 (Fig. 6A). Control animals immunized with PBS lost on average 17.8% of their body weight over the week following SARS-CoV-2 challenge (Fig. 6C). In contrast, animals that were immunized with 10 µg or 2 μg of the bivalent mRNA-VLP vaccine or 10 μg of the bivalent mRNA-native vaccine were protected from weight loss (Fig. 6C). These data demonstrate that immunization with the mRNA-VLP vaccine at the 10 μg and the lower 2 μg dose was sufficient to protect hamsters from weight loss associated with SARS-CoV-2 infection.
Following vaccination and virus challenge, viral load in the oral cavity of hamsters was measured 2 days post virus challenge. Animals that received the control PBS vaccine had geometric mean viral subgenomic RNA (sgRNA) levels of 3.33 × 105 copies/mL at day 2 post virus challenge (Fig. 6D). Hamsters vaccinated with 10 μg of bivalent mRNA-native vaccine or 10 μg of bivalent mRNA-VLP vaccine had a statistically significant reduction in geometric mean viral sgRNA compared to control with measurements of 5.64 × 104 (p = 0.0293) and 2.65 × 104 (p = 0.0003) copies/mL, respectively (Fig. 6D).
Importantly, prevention of virus replication in the lungs of infected animals was observed among vaccinated animals as measured by recovery of infectious virus and viral sgRNA following virus challenge. Animals that received the PBS control had geometric mean median tissue culture infectious dose (TCID50) levels of 1.16 × 109 TCID50/g at day 3 post virus challenge (Fig. 6E). In comparison to PBS control, hamsters vaccinated with 10 μg of bivalent mRNA-native, 10 μg of bivalent mRNA-VLP, or 2 μg of bivalent mRNA-VLP vaccines all had statistically significant reductions (p < 0.01) in viral replication with measurements of ≤1.92 × 104, ≤1.92 × 104, and 6.40 × 104 TCID50/g, respectively (Fig. 6E). By day 7 post virus challenge, the PBS control and vaccinated animal groups had comparable levels of residual virus with geometric mean TCID50 levels ranging between 1.39–3.15 × 104 TCID50/g (p > 0.92) (Fig. 6E). The detection of viral sgRNA is a more sensitive measure of virus replication and was employed to further demonstrate the protection afforded by vaccination. The geometric mean viral sgRNA levels in lung homogenate from animals that received the control PBS were 5.46 × 1010 copies/g at day 3 post challenge (Fig. 6F). However, hamsters immunized with 10 μg of bivalent mRNA-native or 10 μg of bivalent mRNA-VLP vaccines showed decreased levels of viral sgRNA with measurements of 5.75 × 105 (p = 0.0852) and 2.61 × 103 (p < 0.0001) copies/g, respectively, in comparison to PBS control animals (Fig. 6F). By day 7 post virus challenge, the PBS control group had a mean viral sgRNA level of 2.85 × 107 copies/g (Fig. 6F). Except for one hamster in the 10 μg bivalent mRNA-native vaccine group and one hamster in the 2 μg bivalent mRNA-VLP vaccine group, all immunized animals had no detectable viral sgRNA at day 7 (Fig. 6F). Taken together, these results demonstrate the protective benefit of the bivalent mRNA-VLP vaccine at low dosage in preventing SARS-CoV-2 replication in the lungs.
The lungs of SARS-CoV-2 infected hamsters were also evaluated for inflammation and alveolar damage using haematoxylin and eosin (H&E)-stained sections and for the presence of the viral nucleocapsid protein (NP) by immunohistochemistry (IHC). Pathology scores were assigned by a board-certified veterinary pathologist on a scale of 0 to 5, with 0 being normal and 5 being the most severe across different categories of pathology lesions most associated with SARS-CoV-2 infection including: inflammation, type 2 pneumocyte hyperplasia, fibrin deposition or hemorrhage, endothelialitis, and necrosis. Lesions observed on day 3 post virus challenge were predominantly associated with inflammation, fibrin/hemorrhage, endothelialitis, and necrosis (Fig. 7A, B). Lesions observed on day 7 post virus challenge were predominantly associated with inflammation and type 2 pneumocyte hyperplasia (Fig. 7A, B). On day 3 post virus challenge, control animals given PBS had a mean pathology score of 8.9 (Fig. 7B). In contrast, animals immunized with 10 μg of bivalent mRNA-native vaccine or 10 μg of bivalent mRNA-VLP vaccine had mean pathology scores of 0 while those animals immunized with 2 μg of bivalent mRNA-VLP vaccine had a mean pathology score of 0.88 (Fig. 7B). On day 7 post virus challenge, PBS control animals had a mean pathology score of 7.63 while all vaccinated animals with 10 μg of either mRNA vaccine had no pathology score and the group vaccinated with 2 μg of bivalent mRNA-VLP vaccine had a mean pathology score of 0.13 (Fig. 7B). Microscopic findings in the 2 μg bivalent mRNA-VLP vaccine group were limited to small, very infrequent focal changes in individual animals for both day 3 and day 7 post virus challenge. The data demonstrate that both bivalent mRNA-native and bivalent mRNA-VLP vaccines protect hamsters from SARS-CoV-2 induced alveolar damage and inflammation.
A Representative images of lung sections taken from hamsters euthanized on day 3 and day 7 post virus challenge taken at low/×0.6, ×4, ×10, and ×20 magnification from H&E-stained slides. B Lung pathology scores were assigned by a board-certified veterinary pathologist across categories of pathology lesions most frequently associated with SARS-CoV-2 infection: inflammation, type 2 pneumocyte hyperplasia, fibrin/hemorrhage, endothelialitis, and necrosis based on readings of H&E-stained lung sections from SARS-CoV-2 infected hamsters euthanized on day 3 or day 7 post virus challenge. C Representative images of lung sections taken from hamsters euthanized on day 3 and day 7 post virus challenge taken at low/×0.6 magnification from slides stained by immunohistocytochemistry with an antibody specific for SARS-CoV-2 nucleocapsid protein (NP). Brown staining indicates NP-positive foci detected in these lung sections. D The percentage of cells that stained positive for NP from lung sections taken on day 3 post virus challenge. H&E hematoxylin and eosin staining, PBS phosphate-buffered saline, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, VLP virus-like particle.
The lungs of SARS-CoV-2-infected hamsters were further evaluated for the presence of viral NP antigen by IHC. SARS-CoV-2 infection is evident from NP staining in the lungs, especially of the bronchiole and bronchiolar epithelia (Fig. 7C). Consistent with viral load measurements in the lungs (Fig. 6E, F), the NP positivity was noticeably more abundant on day 3 post virus challenge compared to day 7 (Fig. 7C). At day 3 post virus challenge, PBS control animals had NP positivity in an average of 49.1% of lung parenchyma cells (Fig. 7D). In contrast, all immunized animal groups had NP positivity limited to an average of 0.009–0.34% of lung parenchyma cells (p < 0.0001 compared to PBS control; Fig. 7D). These results further demonstrate that immunization with the bivalent mRNA-VLP vaccine reduced SARS-CoV-2 viral load in the lungs of infected hamsters.
Collectively, these results demonstrate that a bivalent mRNA-VLP vaccine protects hamsters against SARS-CoV-2 challenge. Immunization with just 2 μg of bivalent mRNA-VLP vaccine resulted in statistically significant reductions in viral titers in the lungs along with protection from weight loss, pulmonary inflammation, and alveolar damage induced by SARS-CoV-2 infection.
Discussion
mRNA-VLP vaccines are designed to elicit more potent and durable nAb responses with increased breadth against viral variants. Structural analysis of the VLPs as expressed by the mRNA-VLP Delta vaccine confirmed that the spike-ferritin VLPs mimic the spike found on the SARS-CoV-2 virus particle. These conformations are consistent with literature reports of spike (Delta) resolved in isolation and on the virion21 confirming that the spike antigen is appropriately presented as an antigen from the mRNA-VLP vaccine.
Our data showed that VLP presentation of the prototypical Delta spike antigen expressed from an mRNA-VLP vaccine elicited a more potent and broader nAb response in naive mice compared to the native Delta spike antigen expressed from an mRNA-native vaccine. To confirm these findings and interrogate durability of the humoral response, we administered the mRNA-VLP Delta or mRNA-native Delta vaccines to NHP. The mRNA-VLP vaccine elicited nAbs at levels significantly higher for all tested reporter variant viruses compared with the mRNA-native Delta vaccine. In addition, the increased virus nAb levels induced by the mRNA-VLP vaccine were maintained for at least 6.5 months after the first immunization. Consistent with this finding, the mRNA-VLP Delta vaccine induced a trend toward higher levels of spike-specific B cells measured in the periphery shortly after the second immunization and a significant increase in long-lived antibody secreting cells in the bone marrow (BM) at day 196 when compared to the mRNA-native Delta vaccine. Importantly, an exclusive Th1-biased CD4+ T cell response was measured for both mRNA-VLP and mRNA-native vaccines like the approved COVID-19 mRNA vaccines19.
Vaccine potency and efficacy were demonstrated in preclinical models with spike antigen sequences used in an approved bivalent vaccine composition. Such bivalent mRNA-VLP vaccines showed either significantly better nAb responses or a trend toward improved antibody responses against several of the related Omicron pseudoviruses. Furthermore, in the SARS-CoV-2 hamster model, the bivalent mRNA-VLP vaccine showed improved immunogenicity when compared with the bivalent mRNA-native vaccine while also providing protection against SARS-CoV-2-associated weight loss, viral replication in the lungs, as well as alveolar damage and inflammation in the lungs post virus challenge.
The mechanism(s) driving two notable properties of the mRNA-VLP vaccines—increased immunogenicity against viral variants not encoded by the vaccine and the potential for increased durability as observed in NHP—is not clear. The simplest explanation is that the increased vaccine potency drives improved virus nAb responses across all variants and drives higher numbers of long-lived antibody secreting cells from the BM (of NHP). Alternatively, the mRNA-VLP vaccines may drive fundamentally different antibody responses, consistent with those observed for protein-based VLP vaccines in mice22. Additional experiments will be required to distinguish between these possibilities. The higher antigen valency of VLPs provides greater potential for B cell activation10 allowing VLPs to activate a wider range of individual B cells to aid in the potency and/or breadth of the resulting antibody response23. Further aiding in antibody affinity maturation is the observation that VLPs display increased trafficking to draining lymph nodes24.
Several different approaches to generate multivalent vaccines have proven quite successful as protein-based modalities25. However, merging these antigen design concepts with the speed, fidelity, and manufacturability of mRNA allows for the rapid response to highly dynamic pathogens like SARS-CoV-2 and influenza with the potential to generate enhanced immune responses.
The increased potency of the mRNA-VLP vaccines has the potential to enable lower doses in the clinic. In the preclinical hamster model, a fivefold lower dose of bivalent mRNA-VLP vaccine elicited a 15-fold higher level of virus nAb titers against Omicron BA.4/5 compared to the bivalent mRNA-native vaccine (Fig. 6B) and showed comparable protection against challenge with an ancestral SARS-CoV-2 virus. A subsequent Phase I human study (ARTEMIS-C; NCT06147063) of an mRNA-VLP vaccine demonstrated a similar immunogenicity to a currently approved mRNA vaccine while being administered at a lower dosage26. The use of lower doses would reduce the cost of goods with the potential to improve reactogenicity to mRNA/LNP vaccines. Furthermore, lower mRNA vaccine doses may better enable combination vaccines without loss of vaccine potency.
Overall, these studies show that mRNA-VLP vaccines elicit more potent and broader virus nAb responses across several preclinical animal models when compared to vaccines designed to mimic currently approved mRNA vaccines expressing native viral antigens. In addition, the mRNA-VLP vaccines have the potential for improved durability compared to current COVID-19 mRNA vaccines.
Methods
VLP antigen design, DNA plasmid construction, RNA synthesis, and LNP formulation
mRNA-VLP constructs were designed to express spike sequences (retrieved from GISAID) engineered to incorporate the S2P stabilizing substitutions (at positions K986 and V987), ablate the furin cleavage site (RRAS to GSAS), optimize the hinge region of the ectodomain stalk27, and truncate spike at position 1163 to allow an in-frame fusion with a short GSGGSG (glycine-serine) linker followed by the H. pylori ferritin coding sequence as described in Joyce et al.28. Such antigen-coding cassettes were built into an mRNA production vector through standard molecular biology techniques and final constructs confirmed by Sanger sequencing. Linearized DNA templates were used in a run-off T7 RNA polymerase in vitro transcription reaction (New England Biolabs, Ipswich, MA, USA) with the addition of CleanCap AG (TriLink BioTechnologies, San Diego, CA, USA) for co-transcriptional capping and the addition of N1-methylpseudouridine (TriLink BioTechnologies). The purified mRNAs were encapsulated into LNPs employing a multi-step process which involves mixing of the aqueous mRNA and an organic phase of the lipid components (a proprietary cationic lipid, a saturated phospholipid, a PEG-lipid, and cholesterol) of the LNP. After LNP formation, the organic phase is removed, a buffer exchange is performed, sucrose is added, and the LNPs are diluted to the mRNA target concentration and aseptically filtered to generate the bulk mRNA-LNP dispersion. The mRNA-LNP is tested for critical attributes and stored at ≤−65 °C before final use. The LNP systems and accompanying process were developed by and licensed from Acuitas Therapeutics (Vancouver, BC, Canada).
Biophysical characterization
Spike-ferritin VLPs were expressed and purified from Freestyle 293X cells (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, VLP-containing cell culture supernatant was collected and VLPs captured onto an affinity column packed with CNBr-activated Sepharose 4B resin (Sigma Aldrich, St Louis, MO, USA) coupled to an anti-spike monoclonal antibody (AstraZeneca). The affinity column was washed with PBS pH 7.4 and protein eluted with 0.1 M glycine-HCL, 500 mM NaCl pH 2.6. The peak fractions containing protein were pooled and neutralized with 5% v/v 1 M Tris pH 9. The material was dialyzed in PBS and concentrated with a Vivaspin 10 kDa molecular weight cutoff reagent (Sigma Aldrich) according to manufacturer’s protocol. Concentration was determined by measuring absorbance at 280 nm. Purity and mass were determined by SEC-MALS (size exclusion chromatography—multi-angle light scattering) with a Superdex200 10/300 GL column placed in-line with a multi-angle light scattering instrument (Wyatt Technologies, Santa Barbara, CA, USA). Protein size was determined by multi-angle dynamic light scattering (Malvern Panalytical, Malvern, UK) and protein characterized by SDS-PAGE.
Cryo-electron (cryo-EM) microscopy and structure determination
Samples of assembled spike-ferritin VLPs were aliquoted into 10 µl aliquots at 0.7 –0.9 mg/mL frozen at −80 °C and sent to the AstraZeneca Cryo-EM facility in Cambridge, UK. Quantifoil Cu R2/2 200 mesh grids were glow discharged at 30 mA for 30 sec using a PELCO EasiGlow glow discharger prior to use. A volume of 2.5 µl of undiluted sample was applied to each grid and blotted for 3–3.5 sec with a blot force of −3.0 before being plunge-frozen in liquid ethane using a Vitrobot MkIV operating at 100% humidity and 4 °C. Data were recorded on a Titan Krios G4 (Thermo Fisher Scientific) operating at 300 kV equipped with a bottom-mounted Falcon4i direct-detector at a nominal magnification of ×75,000 corresponding to a calibrated pixel size of 1.1 Å/pix. A total of 9360 movies were recorded in electron-event representation mode with a dose rate of 6.34 e−/pix/s for 8.14 sec corresponding to a total dose of 42.77e−/Å2. Movies were recorded over a defocus range of −0.8 µm to −2.4 µm.
Movies were motion corrected in Relion 3.1 using the relion implementation of motion correction, followed by estimation of contrast transfer function parameters using Gct v1.18. A subset of ~1000 VLPs centered on the ferritin cage was manually picked and 2D classes were generated to provide templates for subsequent auto-picking using the relion auto-picking tool. An initial stack of 612,537 particles was picked, extracted (binned by 4), and subjected to two rounds of reference-free 2D classification in CryoSPARC v3.3. Particles that aligned to well-defined classes were selected and imported back into Relion3.1 for 3D classification, searching for 10 classes using an initial model generated using the ab-initio generation in CryoSPARC v3.3. Two rounds of 3D classification were performed: one round of 25 iterations with an angular sampling of 7.5° and a second round of 25 iterations with 1.8° angular sampling and local searches. 156,044 particles belonging to classes showing clear density for ferritin and spike were selected and re-extracted into an 800 pixel box (unbinned). Particles were then refined using 3D-auto refine with a mask around the ferritin core, applying octahedral symmetry to produce a consensus map at 2.2 Å with low-resolution density for spike.
In order to further classify the spike protein, particles were symmetry expanded (with duplicates removed) and re-extracted into a 400pix box centered on the low-resolution density at one 3-fold axis corresponding to the spike. Particles were then imported into CryoSPARC for all subsequent steps. One round of 2D classification was performed to identify particles accurately re-centered and showing clear density for the spike. 311,715 particles were selected, an ab-initio model was generated, and refined without imposing symmetry to produce a consensus map for spike. The particles were then subjected to 3D variability analysis using a mask around the whole spike to identify the conformational variability and were separated into 10 clusters. Clusters corresponding to closed spike, 1 RBD up, and 2 RBDs up were pooled into separate bins and refined using non-uniform refinement to get final reconstructions for the three species. Reported resolutions are based on the gold-standard Fourier shell correlation with a cut-off criterion at 0.14329.
Animal welfare
All murine studies were approved and conducted in accordance with AstraZeneca’s Institutional Animal Care and Use Committee and performed in adherence with the following standards of the AAALAC: the eighth edition of the Guide for the Care and Use of Laboratory Animals, the Animal Welfare Act, and the 2015 reprint of the Public Health Service Policy on Humane Care and Use of Laboratory Animals. The NHP and hamster studies were approved and conducted in accordance with Bioqual’s Institutional Animal Care and Use Committee and performed in adherence with the standards of the AAALAC. Interventions to reduce pain, suffering, and distress were taken whenever possible in accordance with these protocols. The total number of animals used, group sizes, and number of groups were considered the minimum required to properly characterize the effects of the test articles and PBS control. The studies were designed so that they did not need an unnecessary number of animals to accomplish their objectives, with animals randomly assigned to their respective treatment groups.
Immunogenicity in mice
Seronegative BALB/c female mice, 6–8 weeks of age (Inotiv, Lafayette, IN, USA), were separated into groups of 6 mice each and immunized twice IM (left gracilis muscle; 50 μL volume) with 1 μg of LNP-formulated mRNA vaccines. nAb titers against a panel of reporter SARS-CoV-2 pseudoviruses were measured 2 weeks after the second immunization. To establish a cohort of COVID vaccine-primed mice, seronegative BALB/c female mice, 6–8 weeks of age (Inotiv), were IM immunized twice (left gracilis muscle; 50 μL volume) on study days 1 and 21 with 0.2 μg of LNP-formulated mRNA vaccine expressing the native ancestral (Wuhan_D614G) spike antigen. Animals were then rested for ~6.5 months, at which point mice were administered a booster dose of 0.2 μg of LNP-formulated mRNA vaccine as indicated via IM vaccination (left gracilis muscle; 50 μL volume). nAb titers against a panel of reporter pseudoviruses were measured one day prior to and 2 weeks after the booster administration. Anesthesia was not employed for these procedures per institutional IACUC protocol.
SARS-CoV-2 pseudovirus neutralization assay
A third-generation human immunodeficiency virus based lentiviral vector system was implemented to generate SARS-CoV-2 pseudoviruses. Briefly, Freestyle 293X cells (Thermo Fisher Scientific) were co-transfected with the following four plasmids (1): pAZRev (lentiviral packaging plasmid) (2), pPACKH1-Gag (lentiviral packaging plasmid) (3), pESRC-CMV-Luc2p-EF1Puro (luciferase reporter plasmid), and (4) pCAGG-Sdl19Gen (plasmid expressing the SARS-CoV-2 spike protein with 19 amino acid C-terminal deletion). At 48 hours post transfection, the cell supernatant was harvested, cellular debris removed by low-speed centrifugation, and the clarified supernatant passed through a 0.45 µM filter unit. To prepare pseudoviruses bearing the spike from the ancestral Wuhan_D614G or Delta variants, the clarified supernatants were concentrated 100-fold by ultracentrifugation at 10,000 × g for 4 hours at 4 °C. To prepare pseudoviruses bearing the spikes from Omicron BA.1, BA.2, BA.4/5, or BQ.1.1 viruses, the clarified supernatants were loaded on top of a 10% sucrose cushion at 4:1 v/v ratio of virus:sucrose and centrifuged at 10,000 × g for 4 hours at 4 °C.
Serial dilutions of animal sera were prepared in 96-well microtiter plates and pre-incubated with pseudovirus for 60 minutes at 37 °C to which Ad293 cells stably expressing human ACE2 (AstraZeneca) were added. The plates were then incubated for 48 hours at 37 °C whereupon luciferase activity was measured on an EnVision 2105 Multimode Plate Reader (Perkin Elmer, Waltham, MA, USA) using the BrightGlo™ Luciferase Assay System (Promega, Madison, WI, USA) according to the manufacturer’s recommendations. Percent inhibition was calculated relative to pseudovirus alone control.
Immunogenicity in NHP
Immunization of SARS-CoV-2 seronegative NHP was performed at BIOQUAL, Inc. (Rockville, MD, USA). Briefly, 12 cynomolgus macaques (Macaca fascicularis), 29–56 months of age (BIOQUAL, Inc.), were separated into two groups of 6 animals and intramuscularly (IM) immunized twice (quadriceps muscle) on study days 1 and 28 with 10 µg of LNP-formulated mRNA/LNP vaccine in 1 mL. The mRNA-native group was comprised of 2 females and 4 males while the mRNA-VLP group consisted of 1 female and 5 males. Sera were obtained throughout the study to evaluate antibody levels. Whole blood was collected at different timepoints and peripheral blood mononuclear cells (PBMCs) were isolated to evaluate spike-specific T and B cell responses. Bone marrow (BM) aspirates were also obtained using standard bone biopsy procedures following topical anesthetic (bupivacaine) application to the biopsy site. Prior to immunization and sample collection, animals were anesthetized with ketamine HCL (10 mg/kg) administered intramuscularly.
ICS of spike-specific T cells
For the ICS assay, NHP PBMCs were thawed and rested overnight before stimulation with pools of 15-mer peptides overlapping by 10 amino acids covering the SARS-CoV-2 spike protein (JPT Technologies, Berlin, Germany) for 6 hours at 37 °C. After stimulation, cells were washed with PBS and stained with viability dye for 20 min at room temperature, followed by staining for surface T cell markers for 20 min at room temperature, cell fixation and permeabilization with the BD Cytofix/Cytoperm kit (BD Biosciences, Franklin Lakes, NJ, USA) for 20 min at room temperature, and then followed by staining for intracellular markers for 20 min at room temperature. All antibody staining was performed in the dark. T cell phenotype staining and all pre-fixation washes were performed with fluorescence-activated cell sorting buffer (PBS + 1% fetal bovine serum + 0.02% NaN3) and intracellular staining and post-fixation washes were performed in 1× BD Perm/Wash buffer (BD Biosciences). See Supplemental Table 1 for a complete list of antibodies used.
Upon completion of staining, cells were analyzed on a BD FACSymphony A5 SE flow cytometer (BD Biosciences). Samples were invalidated if <10,000 live CD3+ T cells were collected. On average, ≥280,000 cells were collected for each sample. Data were analyzed using FlowJo 10.6.2 (BD Biosciences). Anomalous “bad” events were separated from “good” events using the PeacoQC algorithm30. “Good events” were used for all downstream gating. Individual CD4+ T cell cytokines were plotted on the y-axis versus CD154+ on the x-axis. Only double positive events were used to determine positive responses. Th1 response was defined by CD4+ T cells producing any combination of IFNγ, IL-2, and TNFα. Th2 response was defined by CD4+ T cells producing any combination of IL-4 and IL-13. All antigen-specific cytokine frequencies are reported after background subtraction of identical gates from the same sample incubated with the dimethyl sulfoxide negative control.
B-cell phenotyping assays
Fluorophore-labeled spike antigen probes were generated by the sequential addition of BUV661 streptavidin (BD Biosciences) to biotinylated Delta spike trimer protein (Acro Biosystems, Newark, DE, USA). Probes were evaluated for an appropriate binding profile prior to experimentation using UltraComp Plus beads (Thermo Fisher Scientific) coated with SARS-CoV-2-specific monoclonal antibodies.
NHP PBMCs were thawed after removal from liquid nitrogen storage using thawsomes (Medax International, Salt Lake City, UT, USA) and immediately stained with Live/Dead Blue (Thermo Fisher Scientific) for 15 min at room temperature prior to surface staining for B cell associated surface markers in FACS Buffer supplemented with Brilliant Stain Buffer Plus (BD Biosciences) for 20–30 min on ice. B-cell specificity was assessed following PBMC incubation with the Delta spike trimer probe. See Supplemental Table 2 for a complete list of antibodies used. Samples were fixed with 4% paraformaldehyde (Thermo Fisher Scientific) prior to sample acquisition on a Cytek Aurora Spectral Cytometer (Cytek Biosciences, Fremont, CA, USA). Post-acquisition analysis was completed using FlowJo 9 (BD Biosciences) and GraphPad Prism software version 9.0.0.
NHP BM aspirates, derived from the femur, were processed into single cell suspensions using density gradient centrifugation followed by red blood cell removal with ACK lysing buffer (Thermo Fisher Scientific). BM cell concentrations were determined and cells were stored in liquid nitrogen. Cells were thawed from liquid nitrogen storage using thawsomes (Medax International) and added to ELISpot plates (Mabtech, Nacka Strand, Sweden) coated with 2 μg/mL of recombinant Delta spike RBD protein (AstraZeneca) and incubated for 18 hours at 37 °C. Enumeration of antigen-specific antibody-secreting cells was completed according to the manufacturer’s protocol (Mabtech).
Immunogenicity and vaccine efficacy in a hamster model of SARS-CoV-2 challenge
Hamster immunization and challenge studies were performed at BIOQUAL, Inc. (Rockville, MD, USA). Briefly, Golden Syrian hamsters (Inotiv), 6–8 weeks of age, were separated into four groups of 16 animals (8 females and 8 males) and IM immunized twice (quadriceps muscle) on study days 1 and 21 with the indicated mRNA/LNP vaccines at the indicated doses or PBS control in 0.1 mL volume. nAb titers against a panel of SARS-CoV-2 pseudoviruses were measured 2 weeks after the second immunization (day 35). On study day 42, animals were challenged via intranasal route with 6 × 103 PFU of SARS-CoV-2 strain USA-WA1/2020 (BEI Resources, Manassas, VA, USA) in 50 μL. Post virus challenge, clinical observations were recorded and body weights measured once daily until termination. Oral swabs were collected on study day 44 from all animals. On study days 45 (day 3 post virus challenge) and 49 (day 7 post virus challenge), animals were euthanized by exsanguination followed by thoracotomy and necropsied for lung and nasal turbinate toward viral load analyses and histopathology.
Quantitation of viral sgRNA by quantitative real-time polymerase chain reaction (qRT-PCR)
Quantification of viral sgRNA was performed by the Immunology & Virology Quality Assessment Center/Human Vaccine Institute/Duke University, with a standard methodology. Briefly, a QIAsymphony SP (QIAGEN, Germantown, MD, USA) automated sample preparation platform, along with a virus/pathogen DSP midi kit and the complex800 protocol, was used to extract viral RNA from 800 μL of lung homogenate samples or oral swab samples. SARS-CoV-2 N gene sgRNA was measured by a one-step qRT-PCR as described previously (Li et al. 2021). Briefly, a standard curve for the N gene sgRNA sequence includes the 5’UTR leader sequence, transcriptional regulatory sequence, and the first 227 bp of the N gene. For these assays, TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific) and custom primers/probes targeting the N gene sgRNA (forward primer: 5′-CGATCTCTTGTAGATCTGTTCTC-3′; reverse primer: 5′-GGTGAACCAAGACGCAGTAT-3’; probe: 5′FAM-TAACCAGAATGGAGAACGCAGTGGG-BHQ1-3′) were employed. RT-qPCR reactions were carried out on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific) using the program below: reverse transcription at 50 °C for 5 min, initial denaturation at 95 °C for 20 s, then 40 cycles of denaturation-annealing-extension at 95 °C for 15 s and 60 °C for 30 s. Standard curves were used to calculate N sgRNA in copies per mL. The limit of detection (LOD) for this assay is 200 RNA copies per gram of tissue sample or 31 RNA copies per mL of oral swab fluid.
Quantification of infectious SARS-CoV-2 virus by TCID50 assay
Infectious virus titers in lung tissues were determined by TCID50 assay in Vero E6 cells (ATCC, Manassas, VA, USA). SARS-CoV-2 TCID50 viral titration assays were conducted in BSL-3 laboratories at BIOQUAL, Inc. (Rockville, MD, USA) in accordance with Federal and Institutional Biosafety Standards and Regulations. Briefly, Vero E6 cells were seeded at 2.5 × 104 cells per well (96-well plate) in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% Gentamicin and grown overnight to reach 80–100% confluency at 37 °C. Lung homogenates were ten-fold serially diluted in DMEM supplemented with 2% FBS and 1% Gentamicin. 20 μL of dilutions were added to confluent Vero E6 cells in quadruplicates and incubated at 37 °C for 4 days. The cell monolayers were visually inspected for cytopathic effect (CPE). The presence of CPE is recorded as a plus (+), and the absence of CPE is recorded as a minus (−). The TCID50 value is then calculated using the Read-Muench formula. TCID50 were multiplied by the dilution factor to determine the number of TCID50 per mL of lung homogenates. The TCID50 units per gram of lung tissue were calculated based on the weight of the lung section used to generate the homogenate. The LOD for this assay with the day 3 and day 7 post virus challenge samples was 19,231 and 1923 TCID50 units per gram of tissue sample, respectively.
Histopathology and immunohistochemistry
Hamsters were euthanized on study days 45 and 49, corresponding to 3 and 7 days, respectively, post virus challenge. The left lung lobe, designated for histopathology, was removed and fixed in 10% neutral buffered formalin for 72 hours before transfer to 70% ethanol. Tissues were paraffin-embedded, sectioned at 4 μm, and stained with haematoxylin and eosin. Slides were scanned using a Leica Aperio Scanscope AT2 pathology slide scanner (Leica, Wetzlar, Germany) and whole slide images were evaluated by a board-certified veterinary pathologist who was blinded to the treatment groups. Key histopathologic features were individually recorded as separate findings for study days 45 and 49 and graded on a 5-point scale (1 = minimal, 2 = mild, 3 = moderate, 4 = marked, and 5 = severe).
IHC was performed using an automated Leica Bond RX IHC staining platform (Leica). Following antigen retrieval using Bond ER1 Solution (Leica) for 20 min at 100 °C, slides were stained with a rabbit monoclonal antibody specific for SARS-CoV-2 nucleocapsid protein (Sino Biological, Beijing, PRC) at a dilution of 1:20,000 in Ventana Discovery Ab Diluent (Roche, Indianapolis, IN, USA) at room temperature for 30 min. IHC binding was demonstrated using the BOND Polymer Refine detection kit (Leica) according to the manufacturer’s recommendations. Following cover slipping with HistoCore SPECTRA mounting media (Leica), slides were digitally scanned at ×0.6 or ×40 magnification using a Leica Aperio AT2 pathology slide scanner (Leica). ×40 magnification images were loaded into HALO image analysis software v3.4.2986.230 (Indica Labs, Albuquerque, NM, USA). A DenseNet v2 tissue finder classifier was initially used to identify the lung tissue region; glass regions and air spaces in large airways were segmented and excluded from the analysis. Individual cell segmentation was completed with HALO using the Multiplex IHC module (v3.1.4) with nuclear detection using the AI default settings. The SARS-CoV-2 positivity was determined by optical density threshold for the DAB marker and was expressed as percent positive cells per entire lung parenchyma.
Statistical analysis
Half-maximal inhibitory dilution (ID50) values were determined by nonlinear regression analysis using the GraphPad Prism software version 9.0.0. P values are presented in the figures along with descriptive statistics (e.g., geometric mean and geometric standard deviation) as described in the accompanying figure legends.
Data availability
Data underlying the findings described in this manuscript may be obtained in accordance with AstraZeneca’s data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. Data for studies directly listed on Vivli can be requested through Vivli at www.vivli.org. Data for studies not listed on Vivli could be requested through Vivli at https://vivli.org/members/enquiries-about-studies-not-listed-on-the-vivli-platform/. AstraZeneca Vivli member page is also available outlining further details: https://vivli.org/ourmember/astrazeneca/.
References
Markov, P. V. et al. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 21, 361–379 (2023).
Yang, Y. et al. Longitudinal analysis of antibody dynamics in COVID-19 convalescents reveals neutralizing responses up to 16 months after infection. Nat. Microbiol. 7, 423–433 (2022).
Arunachalam, P. S. et al. Durable protection against the SARS-CoV-2 omicron variant is induced by an adjuvanted subunit vaccine. Sci. Transl. Med. 14, eabq4130 (2022).
Tanne, J. H. Covid-19: Pfizer-BioNTech vaccine is rolled out in the US. BMJ 371, m4836 (2020).
Schoenmaker, L. et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 601, 120586 (2021).
Snapper, C. M. Distinct immunologic properties of soluble versus particulate antigens. Front. Immunol. 9, 598 (2018).
Jackson, N. A. C., Kester, K. E., Casimiro, D., Gurunathan, S. & DeRosa, F. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines 5, 11 (2020).
Liu, C. et al. Self-assembling SARS-CoV-2 spike-HBsAg nanoparticles elicit potent and durable neutralizing antibody responses via genetic delivery. NPJ Vaccines 8, 111 (2023).
Xu, Z. et al. In vivo assembly of nanoparticles achieved through synergy of structure-based protein engineering and synthetic DNA generates enhanced adaptive immunity. Adv. Sci. 7, 1902802 (2020).
Kato, Y. et al. Multifaceted effects of antigen valency on B cell response composition and differentiation in vivo. Immunity 53, 548–63.e8 (2020).
Patel, K. G. & Swartz, J. R. Surface functionalization of virus-like particles by direct conjugation using azide-alkyne click chemistry. Bioconjug. Chem. 22, 376–387 (2011).
Grgacic, E. V. L. & Anderson, D. A. Virus-like particles: passport to immune recognition. Methods 40, 60–65 (2006).
Roldão, A., Mellado, M. C., Castilho, L. R., Carrondo, M. J. & Alves, P. M. Virus-like particles in vaccine development. Expert Rev. Vaccines 9, 1149–1176 (2010).
Reutovich, A. A., Srivastava, A. K., Arosio, P. & Bou-Abdallah, F. Ferritin nanocages as efficient nanocarriers and promising platforms for COVID-19 and other vaccines development. Biochim. Biophys. Acta Gen. Subj. 1867, 130288 (2023).
Kanekiyo, M. et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 (2013).
Houser, K. V. et al. Safety and immunogenicity of a ferritin nanoparticle H2 influenza vaccine in healthy adults: a phase 1 trial. Nat. Med. 28, 383–391 (2022).
Widge, A. T. et al. An influenza hemagglutinin stem nanoparticle vaccine induces cross-group 1 neutralizing antibodies in healthy adults. Sci. Transl. Med. 15, eade4790 (2023).
Ke, Z. et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020).
Corbett, K. S. et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 383, 1544–1555 (2020).
Muñoz-Fontela, C. et al. Animal models for COVID-19. Nature 586, 509–515 (2020).
Benton, D. J. et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 588, 327–330 (2020).
Cohen, A. A. et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science 377, eabq0839 (2022).
Ols, S. et al. Multivalent antigen display on nanoparticle immunogens increases B cell clonotype diversity and neutralization breadth to pneumoviruses. Immunity 56, 2425–41.e14 (2023).
Bachmann, M. F. & Jennings, G. T. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 10, 787–796 (2010).
Bachmann, M. F., van Damme, P., Lienert, F. & Schwarz, T. F. Virus-like particles: a versatile and effective vaccine platform. Expert Rev. Vaccines 24, 444–456 (2025).
Oyedele, T. et al. editors. Safety and immunogenicity of a SARS-CoV-2 mRNA virus-like particle vaccine in adults 18 years of age and older in a Phase I randomized clinical trial. Proc. Int. Soc. Vaccines 2024, 21–23 (2024).
Turoňová, B. et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 370, 203–208 (2020).
Joyce M. G. et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. 37, 110143.
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Emmaneel, A. et al. PeacoQC: Peak-based selection of high quality cytometry data. Cytom. A 101, 325–338 (2022).
Acknowledgements
This study was sponsored by AstraZeneca. We thank James Button, Erin Goetzke, Alexandra Kathenes, Michael Lehmann, Phillip Newton, and Raffaello Cimbro for their technical contributions to the work. We thank Acuitas Therapeutics for developing and providing the LNP and formulation work for these studies.
Author information
Authors and Affiliations
Contributions
Conceptualization: J.P.L., P.S., Y-M.L., J.R.F., W.B.; Formal analysis: J.P.L., O.A., H.A., P.S., Y-M.L., J.R.F., W.B.; Investigation: J.P.L., Y.C., T.K., Y.Z., K.R., K.R., D.F., R.R., O.A., V.C.P., N.J., V.P., S.S., A.P., G.P., S.D., H.A., R.C., H.P., Y.-H.C., G.T., M.N., C.K., J.G., P.S.; writing—original draft: J.P.L.; writing—review & editing: J.P.L., O.A., H.A., H.P., G.T., P.S., Y.-M.L., J.R.F., W.B., M.E.; visualization: J.P.L., O.A., H.A.; supervision: J.P.L., H.A., G.T., P.S., Y-M.L., J.R.F., W.B., M.E.; funding acquisition: J.P.L., W.B., M.E.
Corresponding author
Ethics declarations
Competing interests
J.P.L., Y.C., T.K., Y.Z., K.R., K.R., D.F., R.R., O.A., V.C.P., N.J., V.P., S.S., A.P., G.P., S.D., H.A., H.P., Y.-H.C., G.T., M.N., J.G., P.S., J.R.F., M.E. are current employees of AstraZeneca and hold or may hold stock in AstraZeneca. R.C., C.K., Y.-M.L., and W.B. are previous employees of AstraZeneca and hold or may hold stock in AstraZeneca. AstraZeneca has filed patent applications (WO/2024/153794 and US20240277834) related to mRNA-VLP and mRNA vaccines. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Laliberte, J.P., Cai, Y., Kenny, T. et al. Immunogenicity and efficacy of an mRNA vaccine expressing a virus-like particle spike antigen against SARS-CoV-2. npj Vaccines 10, 258 (2025). https://doi.org/10.1038/s41541-025-01303-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41541-025-01303-w