Abstract
Analyzing three-dimensional excitation-emission matrix (3D-EEM) spectra through machine learning models has drawn increasing attention, whereas the reliability of these machine learning models remains unclear due to their “black box” nature. In this study, the convolutional neural network (CNN) for classifying numbers of fluorescent components in 3D-EEM spectra was interpreted by gradient-weighted class activation mapping (Grad-CAM), guided Grad-CAM, and structured attention graphs (SAGs). Results showed that the original CNN classifier with high classification accuracy may make a classification based on misleading attention to the non-fluorescence area in 3D-EEM spectra. By removing Rayleigh scatterings in 3D-EEM spectra and integrating convolutional block attention module (CBAM) in CNN classifiers, the correct attention of the trained CNN classifier with CBAM greatly increased from 17.6% to 57.2%. This work formulated strategies for improving CNN classifiers associated with environmental fields and would provide great help for water determination in both natural and artificial environments.
Similar content being viewed by others
Introduction
Three-dimensional excitation-emission matrix (3D-EEM) fluorescence spectroscopy has been widely applied in characterizing fluorescence substances (e.g., dissolved organic matter (DOM), soluble microbial product (SMP), and extracellular polymeric substances (EPS)) originated from environmental fields. The fluorescent components in water samples (e.g., aromatic amino acids, humic acids, and flavins) can be sensitively detected by the 3D-EEM fluorescence spectroscopy1,2,3. The fluorescence signals measured by the 3D-EEM fluorescence spectroscopy will change depending on both the environmental conditions and the characteristics of these fluorescent components4,5,6. Therefore, qualitative and quantitative tracking of fluorescent components in water samples can be realized by the 3D-EEM fluorescence spectroscopy7. However, the output of 3D-EEM fluorescence spectroscopy (i.e., 3D-EEM spectra) may be difficult to understand directly due to many disturbing noises and overlapped fluorescent signals8,9.
To this end, chemometrics researchers have developed fluorescence decompose methods, such as parallel factor analysis (PARAFAC)10,11, to disengage overlapped fluorescent signals and output corresponding maps of each fluorescent component12. However, due to the time-consuming procedure and high sample requirement, these methods cannot be embedded into online monitoring systems directly4,11. Our previous work proposed a fast fluorescent identification network (FFI-Net) based on the convolutional neural network (CNN)4. The trained FFI-Net could classify the numbers of fluorescent components in a single 3D-EEM spectrum and predict all maps of these fluorescent components in a few seconds4, which is essential to the online analysis.
Unfortunately, compared with natural images in computer vision (e.g., car or bird images), the overlapped fluorescence signals in 3D-EEM spectra cannot be directly analyzed by visual inspection8. Therefore, although the CNN classifiers for classifying the numbers of fluorescent components in 3D-EEM spectra presented robust performance4, it is still unknown whether the CNN classifiers made the correct classification according to the correct regions of 3D-EEM spectra (e.g., regions of fluorescent peaks). Moreover, we do not understand why the CNN classifiers could distinguish the differences in different 3D-EEM spectra because of the “black box” nature of deep learning models13. The CNN classifier cannot say “I don’t know” in ambiguous situations and instead returns the class with the highest probability14. Therefore, the reliability of CNN classifiers for analyzing 3D-EEM spectra should be further investigated.
The interpretability and explainability of CNN models have received great attention in recent years15,16,17. Many CNNs explanation methods for visualizing convolutional layers or disturbing input images have been developed. For example, the gradient-weighted class activation mapping (Grad-CAM) method has been utilized for producing visual explanations of decisions from CNNs18. Ribeiro et al.19 proposed a local interpretable model-agnostic explanations (LIME) method to form an interpretable surrogate model that is locally faithful to the CNN classifier. Moreover, Shitole et al.20 found that images may have multiple relatively localized explanations, and established structured attention graphs (SAGs) to visualize how different combinations of image regions impact the confidence of a classifier. Meanwhile, some strategies for improving the accuracy of CNNs based on the attention mechanism have been developed21,22,23. For instance, Woo et al.21 proposed a Convolutional Block Attention Module (CBAM), which could be integrated into any CNN architecture to emphasize meaningful features along two principal dimensions (i.e., channel and spatial axes) of CNNs. By integrating the CBAM, the performance of CNNs on the multiple benchmarks of object detection was greatly improved. However, due to the great differences between 3D-EEM spectra and natural images (e.g., car or bird images) used in the above methods, the performance of these methods for interpreting and improving the CNN classifiers for 3D-EEM spectra needs to be reevaluated and reconsidered.
Therefore, this work investigates the reliability of CNN classifiers for analyzing 3D-EEM spectra. The raw CNN classifiers are first interpreted by the Grad-CAM and SAGs methods. Then, strategies including modifying the data quality and CNN structure for refocusing and improving the attention of CNN classifiers are proposed. The classification results between raw and modified CNN classifiers are compared to highlight the importance of improving the CNN classifiers for analyzing 3D-EEM spectra. To our best knowledge, it is the first time to interpret CNN classifiers designed for classifying the numbers of fluorescent components in 3D-EEM spectra. This work reveals and improves the misleading attention of CNN classifiers on the 3D-EEM spectra and will help formulate strategies for developing deep learning models for analyzing water samples and make these models more acceptable to non-deep learning users.
Results
High accuracy and misleading attention of raw CNN classifier
The raw CNN classifier was first trained by the raw data of 3D-EEM spectra containing scatter peaks. The training loss of the raw CNN classifier significantly decreased to 0.092 ± 0.013 after 100 training epochs (Supplementary Fig. 1). Meanwhile, the test accuracy of the raw CNN classifier trained by raw data reached 89.8 ± 2.5% (Supplementary Fig. 2). The raw CNN classifier trained by the raw data seems to have an acceptable classification accuracy (Table 1).
However, the visual explanations of Grad-CAM and guided Grad-CAM for the trained CNN classifier revealed that the misleading attention of the CNN classifier governed the classification for the 3D-EEM spectra (Fig. 1). The heatmaps for the first convolutional layer (Conv1) presented a high consistency with the color maps, implying that the Conv1 focused on all values in the 3D-EEM spectra. In the following convolutional layers (Conv2-5), the heatmaps gradually concentrated in a smaller area, which supports the classification decision of the CNN classifier18. These phenomena were consistent with the CNN visualization results for natural images (e.g., car or bird images), where more concrete features are extracted by the last convolutional layer in the CNN classifier24,25.
As shown in Fig. 1a, b, the CNN classifier trained by raw data highlighted the regions outside the fluorescent peaks in 3D-EEM spectra. Moreover, the Rayleigh scatterings in 3D-EEM spectra drew the most attention from the CNN classifier. Similarly, a heatmap shift from fluorescent peaks to the Rayleigh scattering was observed in Fig. 1c. The guided Grad-CAM further provided fine-grained importance in classification images (Fig. 1)18. The misleading attention of Conv2-5 on the 3D-EEM spectra was revealed by the guided Grad-CAM. These results demonstrated that the accurate classification of the CNN classifier may originate from misleading attention on 3D-EEM spectra.
To quantify this phenomenon, all 3D-EEM spectra in the Conv5 were analyzed by the Grad-CAM method, and mathematical indices including Correct Accuracyi, Wrong Accuracyi, and Correct Attentioni were calculated by Eqs. (1–3). Very low Correct Accuracyi (3.8%-32.3%) and Correct Attentioni (3.8–32.3%) proved that the raw CNN classifier trained by the raw data mainly classified the 3D-EEM spectra according to the regions outside the fluorescent peaks, which significantly impaired the reliability of the CNN classifier (Table 2). Meanwhile, the 3D-EEM spectra in Class 2 received the highest Wrong Accuracyi (96.2%), which may be due to the highest complexity of five fluorescent components in these 3D-EEM spectra4.
The classification results from the raw CNN classifier trained by the raw dataset were further analyzed by the SAGs method20, which can decompose 3D-EEM spectra into sub-regions and evaluate the effects of removing a particular patch on the classification confidence. As shown in Fig. 2a, the raw CNN classifier trained by the raw dataset made correct classification according to the patches on two Rayleigh scatterings. Meanwhile, the true confidence for this 3D-EEM spectra decreased from 100% to 0% when the patches near the Rayleigh scatterings were removed. To understand the wrong classification made by the CNN classifier, SAGs results of two wrong classification results are presented in Fig. 2b, c. The region near the Rayleigh scattering and outside the fluorescent peaks supported the wrong classification for 5 fluorescent components in Fig. 2b. Similarly, removing a particular patch outside the fluorescent peaks led to a decrease in false confidence from 98% to 0% in Fig. 2c. The results of SAGs further proved that the CNN classifier may make classifications based on misleading attention to the 3D-EEM spectra.
a a 3D-EEM spectrum containing four fluorescent components was classified as four fluorescent components; b a 3D-EEM spectrum containing three fluorescent components was classified as five fluorescent components. c a 3D-EEM spectrum containing three fluorescent components was classified as five fluorescent components. Three 3D-EEM spectra were randomly selected from the training dataset.
Improving CNN attention by modified 3D-EEM spectra
The misleading attention of the CNN classifier on the Rayleigh scatterings was observed in the former section (Fig. 1). To solve this issue, the Rayleigh scatterings in 3D-EEM spectra were removed (called cut data). Then, the raw CNN classifier was trained by the cut data. The training loss of the CNN classifier decreased to 0.116 ± 0.019 (Supplementary Fig. 3), and the test accuracy increased from 89.8 ± 2.5% to 91.3 ± 1.2% (Supplementary Fig. 4), indicating that the raw CNN classifier trained by the cut data also achieved acceptable performance (Table 1). The heatmaps of Grad-CAM and guided Grad-CAM both highlighted the fluorescent peaks in the same 3D-EEM spectra (Fig. 3) tested for the raw data (Fig. 1). Meanwhile, the total Correct Accuracyi and Correct Attentioni of the raw CNN classifier trained by the cut data increased from 16.8% and 17.6% to 33.7% and 36.4%, respectively (Table 2). A significant decrease of Wrong Accuracyi for the Class 0 and Class 2 was observed, implying that the raw CNN classifier learned more features of the cut 3D-EEM spectra in Class 0 and Class 2. Moreover, the 3D-EEM spectra without Rayleigh scatterings showed more clear fluorescent peaks (Fig. 3) than the raw 3D-EEM spectra (Fig. 1). Because the Rayleigh scattering normally has very large fluorescent signals and will affect the normalization and transformation of 3D-EEM spectra. The fluorescent data in region of Rayleigh scattering were set to zero during the PARAFAC analysis procedure10. As a result, the number of fluorescent components classified by the trained CNN classifier should depend on the fluorescent peaks rather than the information provided by the Rayleigh scattering. By removing the strong misleading signatures of Rayleigh scattering, the raw CNN classifier showed both higher accuracy and more focused attention.
a Class 0: a 3D-EEM spectrum contains three fluorescent components; b Class 1: a 3D-EEM spectrum contains four fluorescent components; c Class 2: a 3D-EEM spectrum contains five fluorescent components. Three 3D-EEM spectra containing 3–5 components were the same as the 3D-EEM spectra utilized in Fig. 2.
The SAGs of two correct classification results showed that the combination of several patches on the fluorescent peaks supported the correct classification (Fig. 4). The removal of the patch outside the fluorescent peaks did not reduce the confidence of the CNN classifier (Fig. 4a), whereas the removal of patches near the fluorescent peaks significantly reduced the classification confidence (Fig. 4b). Overall, removing the Rayleigh scatterings in 3D-EEM spectra successfully refocused the attention of the CNN classifier from scatterings to fluorescent peaks in 3D-EEM spectra to some extent (Table 2).
Improving CNN attention by integrating CBAM
Although removing Rayleigh scatterings in 3D-EEM spectra optimized CNN attention to some extent, the total Correct Accuracyi and Correct Attentioni of the CNN classifier were still unsatisfactory. To further improve CNN attention on key regions (i.e., fluorescent peaks) in 3D-EEM spectra, the CBAM was embedded into the CNN classifier. The CNN classifier with CBAM trained by the cut data also received acceptable training loss (0.103 ± 0.011) (Supplementary Fig. 5) and test accuracy (91.2 ± 1.2%) (Supplementary Fig. 6). According to the results of the Grad-CAM, the CNN classifier with CBAM possessed much higher total Correct Accuracyi (55.5%) and Correct Attentioni (57.2%) (Table 2). These results demonstrated that the spatial and channel-wise attention in CBAM was useful in improving CNN attention on the 3D-EEM spectra. As a result, the CBAM enhanced CNN classifier focused on target fluorescent area more properly than the raw CNN classifiers.
The attention mechanism provided by the CBAM not only distinguishes important regions but also improves the representation of interests26,27,28. As a result, the applications of CBAM for natural images have also been proven to cover target regions better than the original CNN21. Compared with natural images, 3D-EEM spectra showed great differences, where overlapped signals cover the fluorescent peaks, and no clear semantic meanings can be observed by users. For classifying numbers of fluorescent components, the CBAM refocused the CNN attention from meaningless regions to whole fluorescent regions, increasing the reliability of the CNN classifier on this task.
Discussion
Although deep learning methods for water samples have been increasingly investigated and applied29,30,31, most of them only considered the model accuracy (e.g., classification accuracy) and ignored the risk behind the “black box”. For example, high classification accuracy for microbeads in wastewater (89%)29 and morphology of activated sludge (95%)30 was obtained, whereas no cue can ensure that the CNN classifiers in their study truly extracted the features of microbeads and sludge morphology from training images. The problem of misleading attention found in this study may occur in other CNN models for image-like data with ambiguous semantics, especially in environmental fields29,30,32,33,34. Therefore, this study focuses on analyzing 3D-EEM spectra of water samples through interpretable CNN classifiers. The misleading attention of CNN classifiers for analyzing 3D-EEM spectra was identified through Grad-CAM and SAGs methods. The misleading attention of 3D-EEM spectra may originate from the features of 3D-EEM data. Unlike natural images with clear semantics, the overlapped fluorescent data in 3D-EEM spectra could not be examined by naked eye. Similar to our previous study4, Yu et al.31 designed a deep convolutional autoencoder to extract feature maps from 3D-EEM spectra. However, the feature maps generated by the deep convolutional autoencoder were not validated by interpretation techniques. Xie et al.35 collected 3D-EEM spectra (351 × 21) of oil samples to establish CNN classification models for oil species. The size of 3D-EEM spectra may affect the attention of CNN, whereas they did not examine the attention of trained CNN models. Yan et al.36 also applied interpretable methods to show the 3D-EEM feature maps of CNN classifier for classifying storage year of Ningxia wolfberry samples. They suggested that the attention of CNN classifier observed on periphery of the fluorescent peaks was caused by more important classification contributions in the weak fluorescent components for the storage year of Ningxia wolfberry samples. This may be a possible reason due to different classification targets (storage year in their study) from our study (numbers of fluorescent components). However, the high classification accuracy in their study may also originate from the misleading attention of CNN classifiers. The data improvement and attention mechanisms strategy also could be utilized to improve the attention of CNN classifiers for the storage year of Ningxia wolfberry samples.
The combination of 3D-EEM spectra and decomposition methods (e.g., PARAFAC method) has been widely applied in many environmental fields. For example, the decomposed 3D-EEM spectra can be utilized as surrogate parameters to monitor the fate of environmental substances, including different organic compounds in wastewater-affected water37, polycyclic aromatic hydrocarbons fractions in combustion particular matter38, DOM in surface water and groundwater5,39. Moreover, the decomposed 3D-EEM spectra can represent key indices in water industries, such as monitoring DOM and disinfection byproduct precursors in drinking water treatment processes40,41, and monitoring microbial activities in wastewater treatment processes42,43. Although 3D-EEM fluorescence spectroscopy is more sensitive, more time-efficient, and less expensive than traditional chromatographic methods38, the decomposition methods for 3D-EEM suffer from time-consuming procedures and strict data requirements10, limiting the online monitoring and analysis of 3D-EEM4. The FFI-Net developed in our previous study4 is promising to decompose the overlapped signals directly and replace time-consuming decomposition methods10, whereas its interpretability and acceptability should be improved. To this end, this study proposed a strategy combining data improvement and attention mechanisms to alleviate the misleading attention of CNN classifiers on 3D-EEM spectra. This study strongly improved the accuracy and reliability of deep learning methods applied to fast analyze the 3D-EEM spectra4 of water samples in different fields.
Meanwhile, there are still some limitations in this study. Only 55.5% total True Accuracyi was achieved by the CNN classifier with CBAM, which means that great enhancement of model attention may be realized by further improving data and model structures. On one hand, transforming 3D-EEM data from array (0-9999) to grey image (0-255) causes the loss of information. A more appropriate image form for 3D-EEM spectra or advanced methods for improving grey images may improve the model’s attention. For example, Shi et al.44 utilized the morphological grayscale reconstruction method to pre-enhance the locations of fluorescent peaks in the grey images of 3D-EEM spectra. On the other hand, the attention mechanism has been embedded in many novel model structures, such as recurrent neural network27 and transformer26. Therefore, a more elegant model structure coupling with a strong attention module may further improve the total True Accuracyi of the classification task for 3D-EEM spectra.
Moreover, the classification labels (i.e., numbers of fluorescent components for each 3D-EEM spectrum) of wastewater samples determined by the PARAFAC method may not represent the true numbers of fluorescent components in the 3D-EEM spectra due to the limitations of this method. The PARAFAC method as a superposition model assumes that all chromophores within the mixture absorb and emit light independently45. However, charge-transfer interactions between chromophores (e.g., humic-like components) in wastewater samples will alter emission properties and impact the calculations of PARAFAC45,46. Nowadays, evaluating and verifying this assumption is difficult for datasets containing wastewater samples46. As a result, the wrong numbers of fluorescent components may pass the model validation of PARAFAC and generate wrong classification labels, which will reduce the quality of training dataset. As a data-driven algorithm, the accuracy and attention of CNN models highly depend on the quality of training dataset. The training dataset prepared by PARAFAC in this study may also be improved by using some more advanced 3D-EEM analysis method (e.g., parallel factor framework-clustering analysis (PFFCA)47, and three-direction resection alternating trilinear decomposition (TDR-ATLD) algorithm48) in future applications. Overall, the results of this study may have important implications for online monitoring and analysis of environmental substances through 3D-EEM spectra. Importantly, this work provided strategies for further improvement of CNN classifiers for 3D-EEM spectra collected from different water fields, making them more robust and acceptable.
Methods
Dataset of 3D-EEM spectra
The 3D-EEM spectra collected from SMP and EPS in biological wastewater treatment systems were used for model development. The water samples of SMP and EPS mainly consist of microbial products-rich substances, such as proteins and humic acids, which can be detected by fluorescence spectroscopy. Due to the high complexity of biological wastewater treatment systems, the fluorescence peaks in 3D-EEM spectra collected from SMP and EPS are commonly overlapped3,49. Therefore, we chose these 3D-EEM spectra as classification targets of CNN classifiers. The SMP samples of anaerobic digestion sludge, anammox sludge, and aerobic sludge were collected from the supernatant in the reactors and were filtered by 0.45 μm membrane before measurement. The EPS samples of anaerobic digestion sludge, anammox sludge, and aerobic sludge were extracted with the cation exchange resin (CER, Amberlite 732, sodium form) method described by Frølund et al.50.
The 3D-EEM spectra of the samples were obtained by a fluorescence spectrometer F-7000 (Hitachi Co., Japan). The excitation (Ex) wavelengths ranged from 200 to 600 nm at a 5 nm-interval and the emission (Em) wavelengths ranged from 200 to 600 nm at a 5 nm scanning step. Excitation and emission slits were both maintained at 5 nm and the scanning speed was 30,000 nm/min. All 3D-EEM spectra were preprocessed to a unified format (Ex = 200–450 nm, Ex interval = 5 nm, Em = 250–500 nm, Em interval = 5 nm) before they were transformed to 3D-EEM images.
The collected 3D-EEM spectra were first analyzed by the PARAFAC method to provide the classification labels (i.e., 3, 4 and 5 fluorescent components denoted as class 0, 1 and 2, respectively). Then, the raw 3D-EEM spectra (excitation/emission wavelength of 200–450 nm/250–500 nm and 5 nm interval) were normalized to 0–255 and transformed as an image format (51 × 51 pixels, PNG file) to form the input images.
In our previous study, the FFI-Net achieved acceptable classification accuracy with a 3D-EEM dataset containing Rayleigh scatterings4. Therefore, the raw 3D-EEM spectra (called raw data) containing Rayleigh scatterings were used to form a training dataset first. Then, to eliminate the impacts of Rayleigh scatterings on the attention of CNN classifiers, the Rayleigh scatterings in 3D-EEM spectra were removed before transforming 3D-EEM spectra into the input images (called cut data).
The number of 3D-EEM spectra for 3, 4, and 5 fluorescent components reached 422, 198, and 266, respectively. The imbalanced number of 3D-EEM spectra for classification labels may cause implicit bias. To reduce this implicit bias, the 3D-EEM spectra for 4 and 5 fluorescent components were duplicated once. As a result, the final classification dataset contained 422, 396, and 532 3D-EEM spectra for three labels, respectively (a total of 1350 samples).
Model development of CNN classifiers
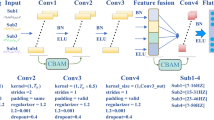
The raw CNN classifier had a similar structure to the famous Alexnet (Fig. 5a)51. Briefly, the CNN classifier first contained five convolutional layers (Conv1-Conv5) and three max-pooling layers, extracting the features from the 3D-EEM spectra. Then, two fully connected layers received the flattened feature map and transformed the information to the output layer with softmax function for three classification labels (i.e., 3, 4 and 5 fluorescent components denoted as class 0, 1 and 2, respectively). To prevent overfitting, the dropout technique was implemented between the last max-pooling layers and the first fully connected layer15. The rectified linear unit (ReLU) was used as the activation function of all convolutional layers and two fully connected layers. The optimizer of the CNN classifier was Adam52 (a common optimizer method in deep learning) with a learning rate of 0.0001, β1 of 0.9, β2 of 0.999, and epsilon of 1 × 10−8.
a The model structure of raw CNN classifier contained five convolutional layers (Conv1-Conv5), three max-pooling layers, two fully connected (FC) layers and one output layer. b Three convolutional block attention modules (CBAM) were embedded into the raw CNN classifier. Two types of input samples (i.e., 3D-EEM spectra with and without Rayleigh scattering) were utilized. c Gradient-weighted class activation mapping (Grad-CAM) method, guided Grad-CAM method, and structured attention graphs (SAGs) method were utilized to interpret the CNN classifier.
To improve the attention of the CNN classifier, the CBAM with both spatial and channel-wise attention was embedded into the CNN classifier (Fig. 5b)21. The CBAM was chosen because it is a lightweight and general module, which can be integrated into any CNN architecture seamlessly with negligible overheads and is end-to-end trainable along with base CNNs21. The channel attention module and spatial attention module were utilized to exploit the inter-channel relationship and inter-spatial relationship of features in 3D-EEM spectra, respectively. The channel attention module containing both average-pooling and max-pooling layers focuses on the meaningful information in the 3D-EEM spectra. Compared with the channel attention module, the spatial attention module focuses on informative regions in the 3D-EEM spectra, which is complementary to channel attention. The average-pooling and max-pooling layers in the spatial attention module were also operated along the channel axis and were applied to generate an efficient feature descriptor21.
The 3D-EEM spectra were resized to 224 × 224 pixels before entering the CNN classifier. The training dataset and test dataset were randomly divided into 80% and 20% for each classification. The performance of CNN classifiers was evaluated by assessing the mean cross-entropy loss and mean classification accuracy. The training loss presented the training performance of the CNN classifiers, whereas the test accuracy showed the classification accuracy of the trained CNN classifiers on unseen data.
Furthermore, to evaluate whether the CNN classifier focused on the fluorescence regions in the 3D-EEM spectra, mathematical indices including Correct Accuracyi, misleading Accuracyi and Correct Attentioni were proposed:
Where i∈{0, 1, 2} represents Class 0 (3D-EEM spectra contain three fluorescent components), Class 1 (3D-EEM spectra contain four fluorescent components), and Class 2 (3D-EEM spectra contain five fluorescent components). The classification performance of CNN classifiers was divided into four types: (I) correct classification with correct attention (CC); (II) correct classification with misleading attention (CM); (III) Wrong classification with correct attention (WC); (IV) Wrong classification with misleading attention (WM). In this way, \({\text{Correct Accuracy}}_{i}\) and \({\text{Wrong Accuracy}}_{i}\) represent correct and misleading extraction features of 3D-EEM spectra by the CNN classifier with correct classification, respectively. \({\text{Correct Attention}}_{i}\) represents the correct attention of the CNN classifier on all 3D-EEM spectra. All raw data and cut data in Conv5 were analyzed by the Grad-CAM method to measure the mathematical indices mentioned above. The final performance results for the CNN classifier are the average of the 6 independent runs.
Interpretation methods for CNN classifiers
The Grad-CAM and guided Grad-CAM were utilized as interpretation methods for the CNN classifier to visualize the attention of convolutional layers on the 3D-EEM spectra (Fig. 5c)18. The Grad-CAM can produce a coarse localization map to highlight the important regions in the 3D-EEM spectra. The guided Grad-CAM method further combined the Grad-CAM visualizations with the guided backpropagation via point-wise multiplication. Compared with Grad-CAM method, high-resolution, highly class-discriminative and more detailed features in the 3D-EEM spectra could be displayed by the guided Grad-CAM method18.
To further interpret the influence of 3D-EEM spectra on the confidence of the CNN classifier, the SAGs method was utilized to visualize how different combinations of image regions (called conjunctions) impact the classifier confidence20. The SAGs method combines multiple saliency maps into a single 3D-EEM spectrum to illustrate multiple different minimal perturbations to change the model output (i.e., the classification result of the CNN classifiers). In this way, the SAGs method could help understand how different combinations of image regions impact the confidence of the CNN classifier. Each 3D-EEM spectrum was divided into 49 = 7 × 7 patches (Fig. 5c) to limit the search space of the Beam Search algorithm. The minimal sufficient explanation (i.e., a minimal region in the image that achieves high classifier confidence) of a 3D-EEM spectrum was presented on the root nodes of SAGs. The SAGs of raw data and cut data were compared to highlight the influence of different 3D-EEM spectra.
It is important to mention that the 3D-EEM spectra are different from the bird or car images tested in the SAGs methods. The latter images have clear identification features, whereas the features of 3D-EEM spectra cannot be directly identified by the naked eyes. Therefore, sufficient conjunctions for classifying 3D-EEM spectra remain to be discovered. Meanwhile, not all 3D-EEM spectra could be interpreted by the SAGs method due to the general high confidence in some 3D-EEM spectra. The interpretation results of the raw CNN classifiers and CNN classifiers with CBAM were compared quantitatively based on the mathematical indices including Correct Accuracyi, misleading Accuracyi and Correct Attentioni (Eqs. 1–3).
Data availability
The data that support the findings of this study are not openly available due to reasons of sensitivity and are available from the corresponding author upon reasonable request.
Code availability
For access to detailed code implementations, please contact the authors directly.
References
Sazawa, K., et al. Effects of paddy irrigation-drainage system on water quality and productivity of small rivers in the Himi region of Toyama, Central Japan. J. Environ. Manage. 342, 118305 (2023).
Sun, Y., Hu, C. & Lyu, L. New sustainable utilization approach of livestock manure: conversion to dual-reaction-center Fenton likecatalyst for water purification. npj Clean Water 5, 53 (2022).
Wang, H.-B. et al. Biofouling characteristics of reverse osmosis membranes by disinfection-residual-bacteria post seven water disinfection techniques. npj Clean Water 6, 24 (2023).
Xu, R.-Z. et al. Fast identification of fluorescent components in three-dimensional excitation-emission matrix fluorescence spectra via deep learning. Chem. Eng. J. 430, 132893 (2021).
Ishii, S. K. L. & Boyer, T. H. Behavior of reoccurring PARAFAC components in fluorescent dissolved organic matter in natural and engineered systems: a critical review. Environ. Sci. Technol. 46, 2006–2017 (2012).
Song, S., Jiang, M., Liu, H., Dai, X., Wang, P. Application of the biogas residue of anaerobic co-digestion of gentamicin mycelial residues and wheat straw as soil amendment: focus on nutrients supply, soil enzyme activities and antibiotic resistance genes. J. Environ. Manage. 335 (2023).
Li, W., et al. A new view into three-dimensional excitation-emission matrix fluorescence spectroscopy for dissolved organic matter. Sci. Total Environ. 855 (2023).
Yamashita, Y. & Jaffe, R. Characterizing the interactions between trace metals and dissolved organic matter using excitation-emission matrix and parallel factor analysis. Environ. Sci. Technol. 42, 7374–7379 (2008).
Luo, J. et al. Simultaneous removal of aromatic pollutants and nitrate at high concentrations by hypersaline denitrification:Long-term continuous experiments investigation. Water Res. 216, 118292 (2022).
Stedmon, C. A. & Bro, R. Characterizing dissolved organic matter fluorescence with parallel factor analysis: a tutorial. Limnol. Oceanogr-Meth. 6, 572–579 (2008).
Murphy, K. R., Stedmon, C. A., Graeber, D. & Bro, R. Fluorescence spectroscopy and multi-way techniques. PARAFAC. Anal. Methods-UK 5, 6557–6566 (2013).
Zeng, X., et al. Recognizing the groundwater related to chronic kidney disease of unknown etiology by humic-like organic matter. npj Clean Water 5, 8 (2022).
Jiménez-Luna, J., Grisoni, F. & Schneider, G. Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2, 573–584 (2020).
Singh, A., Sengupta, S. & Lakshminarayanan, V. Explainable deep learning models in medical image analysis. J. Imaging 6, 52 (2020).
Schramowski, P. et al. Making deep neural networks right for the right scientific reasons by interacting with their explanations. Nat. Mach. Intell. 2, 476–486 (2020).
Talukder, A., Barham, C., Li, X. & Hu, H. Interpretation of deep learning in genomics and epigenomics. Brief. Bioinform. 22, bbaa177 (2021).
Zhang, X. et al. Predicting carbon futures prices based on a new hybrid machine learning: comparative study of carbon prices in different periods. J. Environ. Manage. 346, 118962–118962 (2023).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vision 128, 336–359 (2020).
Ribeiro, M. T., Singh, S., Guestrin, C. & Assoc Comp, M. In “Why Should I Trust You?” Explaining the Predictions of Any Classifier, 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD), pp 1135–1144 (San Francisco, CA, 2016).
Shitole, V., Li, F. X., Kahng, M., Tadepalli, P. & Fern, A. In One Explanation is Not Enough: Structured Attention Graphs for Image Classification, 35th Conference on Neural Information Processing Systems (NeurIPS), Electr Network, Dec 06–14; Electr Network, 34 (2021).
Woo, S. H., Park, J., Lee, J. Y. & Kweon, I. S. In CBAM: Convolutional Block Attention Module, 15th European Conference on Computer Vision (ECCV), 11211 pp 3–19 (Munich, GERMANY, 2018).
Hu, J., Shen, L., Albanie, S., Sun, G. & Wu, E. H. Squeeze-and-excitation networks. IEEE T. Pattern Anal. 42, 2011–2023 (2020).
Wang, Q. et al. In ECA-Net: Efficient Channel Attention for Deep Convolutional Neural Networks, IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 11531–11539 (Seattle, WA, USA, 2020).
Wang, Z. J. et al. CNN explainer: learning convolutional neural networks with interactive visualization. IEEE T. Vis. Comput. Gr 27, 1396–1406 (2021).
Zeiler, M. D. & Fergus, R. In Visualizing and Understanding Convolutional Networks, 13th European Conference on Computer Vision (ECCV), 8689 pp 818–833 (Zurich, SWITZERLAND, 2014).
Vaswani, A. et al. In Attention Is All You Need, 31st Annual Conference on Neural Information Processing Systems (NIPS), 30, (Long Beach, CA, 2017).
Mnih, V., Heess, N., Graves, A. & Kavukcuoglu, K. In Recurrent Models of Visual Attention, 28th Conference on Neural Information Processing Systems (NIPS), 27 (Montreal, CANADA, 2014).
Ren, S., He, K., Girshick, R., Sun, J. & Faster, R.-C. N. N. Towards real-time object detection with region proposal networks. IEEE T. Pattern Anal. 39, 1137–1149 (2017).
Yurtsever, M. & Yurtsever, U. Use of a convolutional neural network for the classification of microbeads in urban wastewater. Chemosphere 216, 271–280 (2019).
Satoh, H., Kashimoto, Y., Takahashi, N. & Tsujimura, T. Deep learning-based morphology classification of activated sludge flocs in wastewater treatment plants. Environ. Sci.: Water Res. Technol. 7, 298–305 (2021).
Yu, J., et al. Detection and identification of organic pollutants in drinking water from fluorescence spectra based on deep learning using convolutional autoencoder. Water. 13, 2633 (2021).
Zhong, S. et al. Machine learning: new ideas and tools in environmental science and engineering. Environ. Sci. Technol. 55, 12741–12754 (2021).
Zhong, S., Zhang, K., Wang, D. & Zhang, H. Shedding light on “Black Box” machine learning models for predicting the reactivity of HO radicals toward organic compounds. Chem. Eng. J. 405, 126627 (2021).
Zhong, S., Hu, J., Yu, X. & Zhang, H. Molecular image-convolutional neural network (CNN) assisted QSAR models for predicting contaminant reactivity toward OH radicals: Transfer learning, data augmentation and model interpretation. Chem. Eng. J. 408, 127998 (2021).
Xie, M., Xu, Q. & Li, Y. Deep or shallow? A comparative analysis on the oil species identification based on excitation-emission matrix and multiple machine learning algorithms. J. Fluoresc. (2023).
Yan, X.-Q., et al. Front-face excitation-emission matrix fluorescence spectroscopy combined with interpretable deep learning for the rapid identification of the storage year of Ningxia wolfberry. Spectroc. Acta Pt. A-Molec. Biomolec. Spectr. 295, 122617 (2023).
Sgroi, M., Roccaro, P., Korshin, G. V. & Vagliasindi, F. G. A. Monitoring the behavior of emerging contaminants in wastewater-impacted rivers based on the use of fluorescence excitation emission matrixes (EEM). Environ. Sci. Technol. 51, 4306–4316 (2017).
Mahamuni, G. et al. Excitation-emission matrix spectroscopy for analysis of chemical composition of combustion generated particulate matter. Environ. Sci. Technol. 54, 8198–8209 (2020).
Schittich, A. R. et al. Investigating fluorescent organic-matter composition as a key predictor for arsenic mobility in groundwater aquifers. Environ. Sci. Technol. 52, 13027–13036 (2018).
Zhang, X. et al. Variations of disinfection byproduct precursors through conventional drinking water treatment processes and a real-time monitoring method. Chemosphere 272, 129930 (2021).
Maqbool, T. et al. Exploring the relative changes in dissolved organic matter for assessing the water quality of full-scale drinking water treatment plants using a fluorescence ratio approach. Water Res. 183, 116125 (2020).
Deng, Y., Li, W., Ruan, W. & Huang, Z. Applying EEM- PARAFAC analysis with quantitative real-time PCR to monitor methanogenic activity of high-solid anaerobic digestion of rice straw. Front. Microbiol. 12, 600126 (2021).
Lourenco, N. D., Lopes, J. A., Almeida, C. F., Sarraguca, M. C. & Pinheiro, H. M. Bioreactor monitoring with spectroscopy and chemometrics: a review. Anal. Bioanal. Chem. 404, 1211–1237 (2012).
Shi, F., et al. Morphological grayscale reconstruction and ATLD for recognition of organic pollutants in drinking water based on fluorescence spectroscopy. Water 11, 1859 (2019).
Sharpless, C. M. & Blough, N. V. The importance of charge-transfer interactions in determining chromophoric dissolved organic matter (CDOM) optical and photochemical properties. Environ. Sci. Process. Impacts 16, 654–671 (2014).
Wünsch, U. J., Murphy, K. R. & Stedmon, C. A. The one-sample PARAFAC approach reveals molecular size distributions of fluorescent components in dissolved organic matter. Environ. Sci. Technol. 51, 11900–11908 (2017).
Qian, C. et al. Fluorescence approach for the determination of fluorescent dissolved organic matter. Anal. Chem. 89, 4264–4271 (2017).
Huang, K., et al. Chemometrics-assisted excitation-emission matrix fluorescence spectroscopy for real-time migration monitoring of multiple polycyclic aromatic hydrocarbons from plastic products to food simulants. Spectroc. Acta Pt. A-Molec.Biomolec. Spectr. 304, 123360 (2024).
Feng, C. et al. Extracellular polymeric substances as paper coating biomaterials derived from anaerobic granular sludge. Environ. Sci. Ecotechnol. 21, 100397 (2024).
Frølund, B., Palmgren, R., Keiding, K. & Nielsen, P. H. Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res. 30, 1749–1758 (1996).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017).
Kingma, D. P., Ba, J. L. Adam: a method for stochastic optimization. In: Proceedings of the 3rd International Conference on Learning Representations (2014).
Acknowledgements
We thank the Fundamental Research Funds for the Central Universities (B240201132), Jiangsu Carbon Peaking and Neutrality Science and Technology Innovation Fund (Grants No. BE2022861), Anhui Provincial Key Laboratory of Environmental Pollution Control and Resource Reuse (2023EPC01) for supporting this work.
Author information
Authors and Affiliations
Contributions
Run-Ze Xu designed the study, analyzed data, drew pictures, and wrote the manuscript; Jia-Shun Cao provided simulation tools and funding; Jing-Yang Luo and Bing-Jie Ni analyzed data and discussed the results; Fang Fang and Weijing Liu provided funding and revised the manuscript. Peifang Wang designed the study, analyzed data and revised the manuscript. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, RZ., Cao, JS., Luo, JY. et al. Attention improvement for data-driven analyzing fluorescence excitation-emission matrix spectra via interpretable attention mechanism. npj Clean Water 7, 73 (2024). https://doi.org/10.1038/s41545-024-00367-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41545-024-00367-w
This article is cited by
-
Effects of Low-Strength Ultrasonic Treatment on Waste Activated Sludge Lysis and Shifts of Microbial Community Traits
Water, Air, & Soil Pollution (2026)
-
An improved CNN model in image classification application on water turbidity
Scientific Reports (2025)
-
Enhanced Tolerance and Growth of Chlorella vulgaris and Scenedesmus quadricauda in Anaerobic Digestate Food Waste Effluent
Applied Biochemistry and Biotechnology (2025)