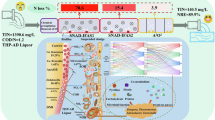
Abstract
Thiocyanate (SCN−) exists in various industries and is detrimental to the ecosystem, necessitating cost-effective and environmentally benign treatment. In response to alleviate the bacterial toxicity of SCN−, this study developed a two-stage coupled system by tandem of anammox in reactor 1 (R1) and SCN−-driven autotrophic denitrification in reactor 2 (R2), achieving simultaneous removal of SCN− and nitrogen. The total nitrogen removal efficiency of the coupled system was 92.42 ± 1.98%, with nearly 100% of SCN− elimination. Thiobacillus was responsible for SCN− degradation. The deduced degradation pathway of SCN− was via the cyanate pathway before coupling, followed by the co-action of cyanate pathway and carbonyl sulfide pathway after coupling. Although scaling-up study is needed to validate its applicability in real-world applications, this study contributes to the advancement of sustainable and cost-effective wastewater treatment technologies, being an attractive path for low-carbon nitrogen removal and greenhouse gas emission-free technology.
Similar content being viewed by others
Introduction
Thiocyanate (SCN−) is commonly found in various industrial processes, including mining, chemical, electroplating, and food processing, especially present at significant levels (300–1500 mg/L) in coking wastewater1,2. However, as one of the top three pollutants in coking wastewater, SCN− often receives less attention and lacks effective control due to its lower toxicity compared to another two pollutants (phenol and cyanide)3. In fact, SCN− exerts a detrimental impact on ecosystem due to its potential contribution as a source of both total nitrogen (TN) and chemical oxygen demand (COD), where theoretically 1 mg SCN− can contribute 0.24 mg N and 1.1 mg COD2. Furthermore, the strong affinity of SCN− for proteins and its inhibitory effect on several enzyme systems-particularly magnesium Adenosine Triphosphatase (Mg-ATPase) pose serious threats to organisms2,4,5. Additionally, the presence of \(-\text{S}-\text{C}\equiv \text{N}\) and \(-\text{N}=\text{C}=\text{S}\) makes SCN− one of the most challenging industrial wastewaters to degrade6. Therefore, it is imperative to seek effective methods for SCN− removal and elucidating degradation pathway to mitigate the harm caused by SCN− in coking wastewater on ecosystem.
Numerous methods are applied to reduce the concentration of SCN− to an environmentally acceptable level ( < 4 mg/L)7. The prevalent physical methods for SCN− removal include coagulation precipitation, membrane extraction, adsorption and ion exchange8. However, the efficiency of these physical methods is often suboptimal and susceptible to variations in environmental parameters, such as pH and nitrate concentration9. For instance, most studies on adsorption reported SCN− removal rates ranging from 90% to 95%10. In addition, the main chemical methods, which encompass electrochemical oxidation, wet air oxidation, ozone catalytic oxidation and chemical oxidation11, are generally more effective than physical ones. Nevertheless, these chemical methods necessitate the expensive equipment and may potentially lead to secondary pollution triggered by the use of additional reagents12. Conversely, biodegradation has emerged as an increasingly prominent approach for SCN− removal, due to its environmental-friendliness and cost-effectiveness in comparison to the inefficient and costly physical and chemical degradation methods9. For instance, Pan et al.13 reported that an autotrophic denitritation process was developed successfully to simultaneously remove thiocyanate and nitrogen. Chen et al.14 observed that autotrophic denitrifying bacteria and anammox bacteria could synergistically remove nitrogen and SCN− by optimizing the NH4+-N/NO2−-N ratio.
To date, a diverse range of bacteria have been identified for their ability to degrade SCN−, including Bacillus15, Pseudomonas16, Alcaligenes17, Thiobacillus18, among others. By action of specific enzymes, the final metabolic products of SCN− are SO42−, CO2 and NH319. Currently, the cyanate (CNO) pathway and the carbonyl sulfide (COS) pathway are two distinct metabolic pathways known for microbial SCN− degradation. In the CNO pathway, the S-C bond of SCN− is cleaved by the action of thiocyanate dehydrogenase (TcDH), resulting in the hydrolysis of SCN− to HCNO and HS− (formula (1)), and HCNO is then hydrolyzed to NH3 and CO2 by the action of cyanase (CYN) (formula (2))9,20. An alternative explanation for the CNO pathway has been put forward, suggesting that SCN− is initially hydrolyzed to CNO− and S2− (formula (3)), followed by the hydrolyzation of CNO− to NH3 and CO2 by the action of CYN (formula (4)) and the oxidation of S2− to S2O32-9,21. In the COS pathway, the N ≡ C bond of SCN− is broken by thiocyanate hydrolase (SCNase), leading to the generation of NH3 and COS (formula (5)). Subsequently, the C-S bond of COS is disrupted by carbonyl sulfide hydrolase (COSase) to produce CO2 and H2S (formula (6))12.
Evidently, both degradation pathways mentioned above necessitate the involvement of oxygen and Wang et al.9 also believed that both CNO and CNS degradation pathways were aerobic.
Under anaerobic or hypoxic conditions, the SCN−-driven autotrophic denitrification where microorganisms can utilize NO3− as a terminal electron acceptor to oxidize SCN−-S, has been proved to be a feasible and effective approach for SCN− removal, utilizing NO3− as electron acceptor. The principle is as formula (7)13,22,23:
Moreover, the SCN−-driven autotrophic denitrification requires no aeration-based external energy input. Furthermore, previous studies have provided supporting evidence that the combination of autotrophic denitrification utilizing reduced-sulfur as electron donor and anammox can be coupled to achieve simultaneous removal of NO3−-N and NH4+-N, thereby reducing energy consumption and sludge production rate24,25,26. Therefore, the two-stage coupled system of anammox and SCN−-driven autotrophic denitrification represents a viable approach for achieving concurrent removal of SCN−, NO3−-N, and NH4+-N from wastewater. Furthermore, anammox and SCN−-driven autotrophic denitrification were operated in separate reactors in the two-stage coupled system, thereby avoiding competition for the intermediate NO2− substrate between anammox bacteria and autotrophic denitrification bacteria. However, the research about SCN− in autotrophic denitrification is limited due to that SCN− is potentially toxic compound, which even at low concentrations may have poisonous effects on microorganisms and reduce the rate of denitrification27. Consequently, investigating the degradation pathway and mechanism of SCN− in autotrophic denitrification and the two-stage coupled system remains the crucial steps towards the efficient removal of SCN− from wastewater.
In this work, anammox in reactor 1 (R1) and SCN−-driven autotrophic denitrification in reactor 2 (R2) were successfully initiated separately and operated stably. Subsequently, the effluent of R1 was used as part of the influent for R2 to construct the two-stage coupled system, which enabled the utilization of the NO3− generated from R1 for participation in the SCN−-driven autotrophic denitrification in R2. As a result, the coupled system effectively accomplished the concurrent elimination of NH4+-N, NO3−-N and SCN−, thereby improving the total nitrogen removal efficiency (TNRE) when compared to that of the solo system. The analysis of microbial structure, the nitrogen and sulfur metabolic pathways, as well as the degradation pathway and mechanism of SCN− before and after coupling were conducted using the 16S rRNA gene amplicon sequencing and metagenomic sequencing data. This research lays a foundation for future practical application of simultaneous removal of SCN− and nitrogen in wastewater.
Results
Acclimation and cultivation of sludge in R1 and R2
The anaerobic granular sludge was first inoculated into two UASB reactors. The influents of R1 and R2 are listed in Supplementary Table 1 and Supplementary Table 2.
As shown in Fig. 1a, influent NH4+-N and NO2−-N in R1were 100 mg/L and 120 mg/L, respectively. After 22 days of operation, both NH4+-N and NO2−-N in effluent were depleted, while the level of NO3−-N remained at 40 mg/L with an accumulation efficiency of 66.70%. According to the equation of anammox (formula (8))28, these data suggested that anammox was successfully initiated in R1, accompanied by a TNRE of 81.80 ± 2.10% and a specific anammox activity (SAA) of 0.51 NH4+-N/g MLSS/h.
Pollutant removal performance of (a) R1 and (b) R2 during the start-up stage. c Analysis of microbial community in R1 at phylum level from 16S rRNA amplicon sequencing data. d The gene abundance of anammox-16S rRNA gene in R1. Analysis of microbial community in R2 at (e) phylum and (f) genus levels from 16S rRNA amplicon sequencing data.
Figure 1b displayed that the concentrations of SCN− and NO3−-N in effluent of R2 exhibited a decreasing trend during 0-20 d, indicating the successful initiation of SCN−-driven autotrophic denitrification. During 20-40 d, there was a significant fluctuation of NO3−-N in effluent as influent NO3−-N and SCN− increased. Subsequently, R2 exhibited stable performance with 85.20 ± 1.54% of TNRE, 0.69 mg SCN−g−1MLSS−1h−1 of specific autotrophic desulfurization-denitrification activity (SADD), and 98.80 ± 0.65% of the removal efficiency for SCN−. In addition, there were also certain amounts of NO2−-N and NH4+-N in effluent, with NH4+-N possibly originating from SCN−-N. The presence of NO2−-N as an intermediate, was attributed to the reaction of denitrification. Furthermore, according to the formula (9), NO2−-N could further oxidize SCN−14, resulting in the low concentration of NO2−-N in effluent.
Analysis of microbial community in R1 and R2
During the operation of reactors (R1 and R2), sludge samples extracted from R1 (R1-50d and R1-80d) and R2 (R2-30d and R2-60d), as well as the initial sludge (IS), were subjected to 16S rRNA gene amplicon sequencing to analyze the microbial community.
Figure 1c exhibited that the microbial community structure in R1 was analyzed at the phylum level, first. The relative abundance of Planctomycetes in sample R1-50d and R1-80d were 4.55% and 4.61%, respectively, apparently higher than 0.94% of initial sludge (IS). To date, a total of six genera of anammox bacteria have been identified across the Earth’s ecosystem, all classified under Planctomycetes29,30,31,32,33. The observed increase in the relative abundance of Planctomycetes may suggest a potential enrichment of anammox bacteria in R1. The anammox-16S rRNA gene analysis was applied to characterize the content of anammox bacteria in R1 sludge and Fig. 1d showed that the abundance of anammox-16S rRNA gene on 60 d increased by 30.57% compared to that of IS, further suggesting that the anammox bacteria was enriched in R1. Even though the microbial community analysis showed a low relative abundance of Planctomycetes, the consistent increase in anammox-16S rRNA gene abundance aligned with the observed TNRE of 81.80 ± 2.10% in R1, indicating that the anammox bacterial community effectively handled the nitrogen load in R1.
As displayed in Fig. 1e, the phylum Proteobacteria occupied the highest relative abundance in sample R2-30d and R2-60d, with percentages of 27.32% and 28.91%, respectively, surpassing the 1.13% in sample IS. Since Proteobacteria contains a large number of denitrifying bacteria34, their increased relative abundance indicated the denitrifying bacteria enriched in R2. In addition, the phylum Bacteroidetes, which is essential for the nitrogen cycle process35, exhibited relative proportions of 35.72%, 24.82% and 29.67% in sample IS, R2-30d, and R2-50d, respectively. The relative abundances displayed a pattern of initial decrease followed by subsequent increase, potentially attributed to the inhibitory effects of toxic SCN− on Bacteroidetes growth during the initial stage of R2 operation. And subsequently, the proliferation and metabolism of Bacteroidetes gradually returned to normal levels due to the enhancement of microbial tolerance.
At the genus level (Fig. 1f), the relative proportion of PHOS-HE36 in sample R2-30d (11.96%) and R2-60d (16.46%) owned an increase compared to that in sample IS (0.25%). PHOS-HE36 played a significant role in both heterotrophic and autotrophic denitrification36, thus also indicating the enrichment of denitrifying bacteria in R2. Similarly, the relative abundances of Thiobacillus in samples R2-30d and R2-60d were 11.11% and 6.81%, respectively, surpassing the absence of Thiobacillus in sample IS. As an obligate autotroph, Thiobacillus gains energy by oxidizing sulfide, thiosulfate, and sulfur to oxidative sulfide through the utilization of oxygen, nitrate, and nitrite as electron acceptors37,38,39. Moreover, Thiobacillus has been found to be the predominant bacterium for degradation of SCN− in aerobic or anoxic conditions40. Therefore, it was considered that Thiobacillus played the dominant role in the removal of SCN− in R2.
Subsequently, the nitrogen and sulfur metabolism in R2, as well as the degradation pathway of SCN− in R2 before coupling, were analyzed based on the short-term batch experiment, 16S rRNA gene amplicon sequencing data and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Nitrogen metabolism in R2 before coupling
Firstly, the short-term batch experiment was performed to explore the transformation path of SCN−-N in R2. Supplementary Fig. 1 showed that no traces of NO2−-N, NO3−-N and SCN−-N were detected at the end of one cycle, while the level of NH4+-N was measured at 52.17 mg/L, which was lower than the initial SCN−-N (77.66 mg/L). It has been demonstrated that NH4+-N is the ultimate degradation product of SCN−-N, which does not serve as an electron donor22,27. Approximately 10% of NH4+-N is transformed into biomass by microorganisms as a nitrogen source20. Considering that anammox is the predominant pathway for NH4+-N removal in anaerobic environment41, and Fig. 1e indicated the presence of Planctomycetes related to anammox in R2, it can be inferred that the annamox process was involved in NH4+-N consumption in R2, despite not obtaining relative abundances of corresponding genes from 16S rRNA gene amplicon sequencing data.
Furthermore, the genes responsible for encoding crucial enzymes involved in the process of nitrogen metabolism were identified and analyzed based on the 16S rRNA gene amplicon sequencing data and KEGG database. As observed in Fig. 2a, the significant increase in relative abundances of functional genes (narGHI, nirS, nirK and nosZ) associated with denitrification indicated that the successful establishment of SCN−-driven autotrophic denitrification in R2 before coupling. While norB exhibited a decreasing trend, it was likely that the presence of SCN− in R2 negatively affected the microbial populations responsible for the production of nitric oxide reductase ([EC: 1.7.2.5]). Meanwhile, glnA (encoding EC: 6.3.1.2) associated with the syntheses of L-Glutamine had a high relative abundance in R2, which suggested NH4+-N was transformed into biomass by microorganisms.
Relative abundances of genes encoding enzymes in (a) nitrogen metabolic pathway and (b) sulfur metabolic pathway in R2 before coupling. The data were referenced from the KEGG database based on the 16S rRNA gene amplicon sequencing data. The relative abundances of genes were determined by normalizing the gene concentrations and comparing them by using targeted reads per million mapped reads (rpmr).
Therefore, the above results indicated that denitrification was responsible for the removal of NO3−-N in R2. Furthermore, NH4+ from SCN−-N degradation was consumed by the transformation into biomass and anammox, resulting in the undetectable level of NH4+-N in R2 effluent (Fig. 1b).
Sulfur metabolism pathway in R2 before coupling
The ultimate degradation product of SCN−-S was SO42− 22,27. In the short-term batch experiment, Supplementary Fig. 2 displayed that 69.40% of SCN−-S was converted to SO42− within one cycle. Similarly, the genes responsible for encoding crucial enzymes involved in sulfur metabolism were identified and analyzed according to the 16S rRNA gene amplicon sequencing data and KEGG database (Fig. 2b). The results revealed varying degrees of increase in relative abundances of genes encoding enzymes responsible for oxidizing S2− to S0 or SO42− during operation of R2. Additionally, there was a significant rise in the relative abundances of genes related to the Sox system. Stratford et al.21 previously reported that a bacterium could oxidize S2− to S2O32−. In R2, there might be microorganisms capable of oxidizing S2− to S2O32−, which subsequently undergone oxidation to SO42− by the sulfur-oxidizing enzyme complex (Sox) system.
The degradation pathway of SCN− in R2 before coupling
Based on the nitrogen and sulfur metabolism (Fig. 2) and the relative abundance of cynS encoding enzyme CYN from 16S rRNA gene amplicon sequencing data, it could be inferred that anaerobic R2 harbored the CNO pathway with NO3− serving as an electron acceptor. Figure 3 exhibited the conjectural degradation pathway of SCN− in R2. Unfortunately, 16S rRNA gene amplicon sequencing data did not provide information regarding the gene encoding TcDH. To date, the gene encoding TcDH has only been reported in the genomes of Thioalkalivibrio species and Thiohalobacter thiocyanaticus FOKN142. Kantor et al.40 found that the complete gene encoding TcDH was related to Thiobacillus, the relative abundances of which in samples R2-30d and R2-60d were 11.11% and 6.81%, respectively. Hence, it can be further indicated that Thiobacillus likely played a dominant role in the degradation of SCN−, especially during the initial step where SCN− was hydrolyzed to HCNO (formula (1)) or CNO− (formula (3)).
Performance of the two-stage coupled system
The two-stage coupled system was constructed successfully by coupling R1 and R2 in series. As shown in Fig. 4a, the removal efficiency of NO2−-N, NH4+-N and SCN− were approximately 100% and no obvious fluctuation was observed in two-stage coupled system, which was attributed to the matured sludge in R1 and R2. And Fig. 4b exhibited that the TNRE, nitrogen removal rate (NRR), and nitrogen loading rate (NLR) were 92.42 ± 1.98%, 0.26 ± 0.04 kg/m3/d, and 0.27 ± 0.04 kg/m3/d, respectively. The TNRE of the two-stage coupled system increased from 85.20 ± 1.54% (solo R2) to 92.42 ± 1.98%. Despite that, the complete removal of NO3−-N was not achieved, and a residual concentration of approximately 10 mg/L NO3−-N was observed in the effluent from R2. Improving the influent concentration of SCN− could be an effective measure to achieve the goal of completing the removal of NO3−-N.
a The concentrations of NO3−-N, NH4+-N, NO2−-N and SCN−; b NLR (nitrogen loading rate), NRR (nitrogen removal rate) and TNRE (total nitrogen removal efficiency). The analysis of microbial community in R2 from metagenomic sequencing data during operation of the two-stage coupled system at (c) phylum level, d genus level and (e) species level.
Analysis of microbial community in R2 after coupling
During the operation of the two-stage coupled system, microbial community compositions of sludge samples collected on day 30, 60 and 90 in R2 were analyzed based on the metagenomic sequencing. As observed in Fig. 4c, the phyla Euryarchaeota and Proteobacteria were frequently detected and their relative abundance kept stable basically in all samples. Figure 4d showed that Methanosaeta, Methanobacterium and Thiobacillus successively were the most abundant genera in R2 sludge after coupling. Under anaerobic conditions, Methanosaeta could maintain the stability of sludge structure and improve the reactor’s ability to resist load fluctuations so that the performance of reactor would not change remarkably43. Though the relative abundance of Thiobacillus exhibited a decline trend (4.37%, 1.73% and 1.58%) and was lower than that of solo R2 (11.11% and 6.81%), Fig. 1b showed that the effluent SCN− concentration was nearly undetected throughout running time (0-90 d). As observed in Fig. 4e, Methanosaeta_harundinacea, a member of genus Methanosaeta, which was conducive to enhancing granular sludge strength and maintaining sludge stability44, occupied the highest proportion in three sludge samples. In addition, Thiobacillus_denitrificans ranked third in relative abundance in all samples. Hao et al.45 reported that Thiobacillus_denitrificans usually existed in the sulfur-driven denitrification processes and could take up S element into cells and transform it. Thus Thiobacillus_denitrificans was regarded as the main species to remove SCN− in R2 after coupling.
In comparison to the solo R2 before coupling, the notable changes of the microbial community composition in R2 after coupling primarily characterized by the emergence of phylum Euryarchaeota and genera Methanosaeta and Methanobacterium, alongside the disappearance of phylum Bacteroidetes and genus PHOS-HE36. The genera Methanosaeta and Methanobacterium both belong to phylum Euryarchaeota and possess the ability for methane production46,47. And the genus Methanosaeta was predominantly represented by the species Methanosaeta_harundinacea and Methanosaeta_concilii, which are also related to methane generation48,49. It can be inferred that a small amount of methanogens was transferred from R1 to R2 along with NO3−-N in effluent of R1. The environmental condition in R2 may provide an optimal habitat for the growth and reproduction of methanogens, leading to the highest relative abundance of Methanosaeta_harundinacea, which possesses the ability to convert CO2 to methane50. The phenomenon was possibly attributed to the presence of CO2, which is the metabolic product of SCN−-C19. In addition, with the prolongation of operation time in R2 (from 30 d to 90 d), it was inferred that Thiobacillus_denitrificans gradually assumed a dominant role in the denitrification process under the influence of SCN−, displacing PHOS-HE36 which had denitrification functionality.
Nitrogen metabolism in R2 after coupling
During the running of the two-stage coupled system, the functional genes and the key enzymes involved in nitrogen metabolism in R2 after coupling were identified and analyzed based on the metagenomic sequencing data and KEGG database. Figure 5a showed that the relative abundances of genes and encoding enzymes associated with denitrification remained relatively stable in R2 before and after coupling, which suggested that a steady and efficient removal of NO3−-N was still primarily via denitrification in R2 during the operation of the two-stage coupled system (30-90 d). As observed in Fig. 5b, the increasing trend in the relative abundances of hydrazine synthase (EC: 1.7.2.7) and hydrazine dehydrogenase (EC: 1.7.2.8), enzymes associated with anammox51, could be attributed to the fact that NO3−-N originated from R1, which was undergoing anammox. As displayed in Fig. 5a, notwithstanding the relatively low abundances of anammox-related genes, it was discernible that anammox contributed to the depletion of NH4+-N in R2 after coupling. Concurrently, the maintenance of stable relative abundances of genes and the enzyme glutamine synthetase (EC: 6.3.1.2), which is related to the synthesis of L-Glutamine52, indicated that a portion of NH4+-N was consumed by the microbial utilization in R2 after coupling, consistent with the situation before coupling.
a The schematic diagram of nitrogen metabolic pathway and the relative abundances of genes encoding key enzymes. b The heatmap of key enzymes associated with nitrogen metabolism. c The schematic diagram of sulfur metabolic pathway and the relative abundances of genes responsible for encoding crucial enzymes. d The relative abundance of module M00595 (S2O32− oxidation by Sox complex). e The heatmap of key enzymes related to sulfur metabolism.
Sulfur metabolic pathway in R2 after coupling
Similarly, the analysis of functional genes and crucial enzymes involved in sulfur metabolism in R2 after coupling was conducted based on the metagenomic sequencing data and the KEGG database. As shown in Fig. 5e, the relative abundances of genes encoding the enzyme sulfide dehydrogenase (EC: 1.8.2.3), responsible for converting S2− to S0 53, decreased compared to those observed in R2 before coupling. Conversely, the relative abundances of genes encoding the enzyme sulfite dehydrogenase (EC: 1.8.5.6), associated with the conversion of SO32− to SO42− 54, significantly increased compared to those in R2 before coupling. As observed in Figs. 5c and 5d, during the operation of the two-stage coupled system (30-90 d), the relative abundances of genes (soxA, soxB, soxX, soxY and soxZ) associated with the Sox system and module M00595 (S2O32− oxidation by Sox complex) exhibited a downward trend in R2. The observed phenomenon implied that the conversion pathway of S2− to SO42− was altered in R2 after coupling compared to that in solo R2, but the underlying reasons for this change was not clear.
The degradation pathway of SCN− in R2 after coupling
According to the nitrogen and sulfur metabolism (Fig. 5) and the relative abundances of genes involved in SCN− degradation, Fig. 6 illustrates the degradation pathway of SCN− in R2 after coupling. In contrast to before coupling, the genes encoding enzyme SCNase (formula (5)) related to the COS pathway (iii) and enzyme CYN (formula (2) and (4)) related to the CNO pathway (i) and (ii) were detected, suggesting a potential coexistence of CNO and COS pathways for SCN− degradation in R2 after coupling. The genes encoding COSase (formula (6)), however, were undetected, consistent with previous studies reporting the absence of any known genes encoding COSase9,55. The decline trend observed in the relative abundance of SCNase, CYN and their associated genes made it difficult to determine which degradation pathway dominated in R2 after coupling. Nevertheless, it was possible that the decrease in relative abundances of Module M00595 (S2O32− oxidation by Sox complex) and its associated genes could be attributed to the appearance of the COS pathway, resulting in reduced S2O32− content. Some scholars believed that the COS pathway was affiliated with Thiobacillus56. Therefore, Thiobacillus still played a crucial role in SCN− degradation in R2, especially Thiobacillus_denitrificans, after coupling during the operation of the two-stage coupled system, despite the possibility that SCN− degradation may occur through the combined action of CNS and COS pathway.
Discussion
Traditional biological nitrogen removal techniques not only require substantial aeration and energy, but also result in huge production of residual sludge. Moreover, these methods necessitate organic carbon sources, which in turn, generate substantial CO2 emissions, exacerbating the climate change crisis and thwarting carbon neutrality. The two-stage coupled system proposed in this study combined anammox with SCN−-driven autotrophic denitrification in series, thus the NO3−-N produced by anammox can be used as electron acceptors for denitrification, further improving TNRE of the system and achieving the simultaneous removal of SCN− and nitrogen. Furthermore, the combination of anammox and autotrophic denitrification makes the two-stage coupled system possess numerous merits: minimal sludge generation, excellent nitrogen removal efficiency, dispensing with the need for aeration and organic carbon sources. Therefore, the two-stage coupled system based on anammox and SCN−-driven autotrophic denitrification, as viable paths for low-carbon nitrogen removal and greenhouse gas emission-free technologies, could represent essential cornerstones of innovation in the field of wastewater treatment towards carbon neutrality.
While the two-stage coupled system offers significant advantages, potential challenges still exist. The biological treatment process requires cultivation and enrichment of specific microbial communities, a task that often demands long time for initiation and stabilization, especially given the slow growth rate of anammox bacteria. Additionally, high concentration of SCN− is toxic to the microbial community, affecting the stability of the system and its ability to efficiently remove contaminants. On the other hand, substantial initial investment is required for constructing and setting up a two-stage coupled system.
In practical applications, a two-stage coupled system can be integrated as a module into existing wastewater treatment plants, especially those that need to treat industrial wastewater containing SCN− and other nitrogen compounds. The seamless integration of the two-stage coupled system with the existing wastewater treatment facilities may involve modifications or expansions of the current infrastructure, as well as ensuring compatibility with the existing processes. In addition, given that the composition of industrial wastewater can vary significantly, it can affect the activity of microorganisms and treatment efficiency. Therefore, long-term monitoring and evaluation of the system are necessary to ensure its continuous effectiveness and adaptability to environmental changes. Two-stage coupled systems have significant potential in wastewater treatment but require comprehensive consideration of technical, economic, and environmental factors to ensure their successful application and sustainable development. Moreover, further exploration and validation of the contribution of two-stage coupled systems in reducing greenhouse gas emissions can be achieved through life cycle assessment, allowing for analysis of carbon footprint of the system at different stages to identify its greatest potential for emission reduction. Thus, a more comprehensive demonstration can be provided on the contribution of two-stage coupled systems in reducing greenhouse gas emissions and mitigating global warming potential, providing scientific basis for future research and practices.
Methods
Start-up and operation of R1 and R2
Firstly, anaerobic granular sludge was inoculated in two UASB reactors with MLSS (mixed liquor suspended solids) in R1 and R2 was 4500 ± 350 mg/L and 4200 ± 420 mg/L, respectively. The influents of R1 and R2 were synthetic wastewater and the components are listed in Supplementary Table 1 and Supplementary Table 2. Specifically, the influent NH4+-N and NO2−-N of R1 were 100 mg/L and 120 mg/L, respectively, during the entire operation. In R2, the initial concentrations of NO3−-N and SCN− in the influent were 80 mg/L and 50 mg/L, respectively for the first 20 days of operation. These concentrations were subsequently rose to 100 mg/L and 75 mg/L. The pH of the influent was adjusted to about 7.5 with 1 M HCl or NaOH solution and the dissolved oxygen (DO) was kept less than 0.5 mg/L by aerating with high-purity argon gas for 10-15 minutes every day. The hydraulic retention time (HRT) of R1 and R2 was both 24 h with continuous water inlet. The water quality was monitored every 3 days and the sludge was regularly sampled for subsequent analysis during operation of R1 and R2.
Construction of the two-stage coupled system
As displayed in Supplementary Fig. 3, the effluent of R1 was used as part of the influent of R2 through a rubber tube and a peristaltic pump after the successful operation of anammox (R1) and sulfide SCN−-driven autotrophic denitrification (R2), respectively. Thus, NO3− generated in R1 could be consumed by the denitrification reaction in R2. The influent was synthetic wastewater and the composition was listed as Supplementary Table 2 and Supplementary Table 3. Specifically, the influent NH4+-N and NO2−-N were 100 mg/L and 120 mg/L, respectively. The HRT of the two-stage coupled system was 24 h with continuous water inlet.
Water quality analysis
The levels of NH4+-N, NO3−-N and NO2−-N were measured with the Standard Methods57. The measurement of SCN− content was conducted utilizing the Ferric nitrate spectrophotometric method as described in previous research22. The level of SO42− was measured by ion chromatography (ICS-6000, Thermo, USA).
Short-term batch experiment
The reaction pathways of N and S from SCN− were explored by short-term batch experiment. Briefly, 10 g fresh sludge was taken from R2 during stable operation and then transferred to a 120 mL serum bottle after being washed 3 times with phosphate-buffered saline (PBS). 100 mL of synthetic wastewater for R2 was added into the serum bottle and regulated the pH of mixture to 7.45 with 1 mol/L HCl or NaOH. The serum bottle was immediately sealed with a butyl rubber stopper after being purged with argon gas for 20 minutes to create an anaerobic environment, subsequently stirred at 180 r/min under 35 °C without light. Under the anaerobic environment in the bottle, 5 mL of water was taken from the bottle every 2 h using a syringe and then subjected to centrifugation for 5 min (5000 r/min). The obtained supernatant was used for the detection of NH4+-N, NO2−-N, NO3−-N, SCN− and SO42−.
The real-time qPCR experiments
The inoculated sludge samples in R1 was named as IS. The sludge samples collected at 30 d and 60 d of R1 operation were named as R1-30d and R1-60d, respectively. DNA was extracted from the sludge samples according to the instruction of PowerSoil® DNA Isolation Kit (MO BIO Laboratories, USA). To ensure the suitability of DNA samples for subsequent experiment, a microspectrophotometer (K5500, USA) was employed to assess their concentration. The target genes analyzed in this work were the anammox-16S rRNA gene, for which the primers used were AMX809F (5’-GCCGTAAACGATGGGCACT-3’) and AMX10666R (5’-AACGTCTCACGACACGAGCTG-3’)58. A MyiQ2 real-time PCR detection instrument (Bio-Rad, USA) was applied to carry out qPCR experiments. The total volume required for conducting this experiment was 20 μL, which included 10 μL of SYBR enzyme, 7.8 μL of RNase-Free Water, 0.4 μL each of forward and reverse primer, 0.4 μL of ROX, and 1 μL DNA extracted from sludge samples. The amplification procedures are shown in Supplementary Table 4. The standard curve for qPCR is presented in Supplementary Fig. 4. All experiments were repeated three times.
16S rRNA gene amplicon sequencing
With the exception of the IS sample, the sludge samples were collected at 50 d and 80 d of R1 operation (before coupling), as well as at 30 d and 60 d of R2 operation (before coupling). These samples were named as R1-50d, R1-80d, R2-30d and R2-60d, respectively. The OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek, Norcross, GA, USA) was utilized for the extraction of the total genomic DNA from the aforementioned sludge samples. The extraction process followed the instructions provided by the manufacturer, and the DNA obtained was subsequently preserved at -20 °C. Subsequently, the quantity was measured with the NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and the quality was assessed utilizing the 1.2% agarose gel electrophoresis.
The PCR amplification of the V3-V4 region in bacterial 16S rRNA genes was performed using a forward primer 515 F (5’-TWNGGCATRTGRCARTC-3’) and a reverse primer 907 R (5’-CCGTCAATTCMTTTRAGTTT-3’) were employed for. To facilitate multiplex sequencing, unique 7-bp barcodes specific were integrated into the primers for each sample. PCR amplicons were subjected to purifying using Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantification was carried out with the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). The amplicons were pooled in equal amounts after quantifying each individual. Subsequently, the pair-end 2×250 bp sequencing was conducted on Illlumina NovaSeq platform with NovaSeq 6000 SP Reagent Kit (500 cycles) at Shanghai Personal Biotechnology Co., Ltd (Shanghai, China).
The microbiome data analysis was conducted using QIIME 2 software59 with minor adjustments based on the official tutorials (https://docs.qiime2.org/2019.4/tutorials/). The taxonomic compositions and abundances were visualized using MEGAN60 and GraPhlAn61 for data analysis. Microbial functions were predicted by PICRUSt2 (Phylogenetic investigation of communities by reconstruction of unobserved states)62 upon MetaCyc (https://metacyc.org/) and KEGG (https://www.kegg.jp/) databases. The raw data have been submitted to the NCBI sequence read archive with accession number SAMN40867343 (IS), SAMN40867344 (R1-50d), SAMN40867345 (R1-80d), SAMN40867346 (R2-30d) and SAMN40867347 (R2-60d).
Metagenomic sequencing
The sludge samples were collected at 30 d, 60 d and 90 d of R2 operation (after coupling) and named as 30d, 60d and 90d, respectively. The total genomic DNA was extracted from the obtained sludge samples with the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) accordance with the instructions provided by the manufacturer. The TBS-380 and NanoDrop2000 were utilized to assess the concentration and purity of the extracted DNA, respectively. Additionally, a 1% agarose gel was employed to evaluate the quality of the extracted DNA.
The extracted DNA sample was fragmented into an average size of around 400 bp using the Covaris M220 (Gene Company Limited, China). Subsequently, the NEXTFLEX Rapid DNA-Seq kit (Bioo Scientific, Austin, TX, USA) was employed to construct the paired-end library.
The Illumina Novaseq 6000 (Illumina Inc., San Diego, CA, USA) was utilized for paired-end sequencing at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) following the manufacture’s guidelines provided in NovaSeq Reagent Kits (www.illumina.com). To guarantee the precision of subsequent data analysis, fast63 (https://github.com/OpenGene/fastp, version 0.20.0) was employed to filter out reads of insufficient quality (length < 50 bp, quality value < 20 or containing N bases). The metagenomics data underwent assemble using MEGAHIT64 (https://github.com/voutcn/megahit, version 1.1.2). Relevant tool, such as Prodigal65 / MetaGene66 (http://metagene.cb.k.u-tokyo.ac.jp/), were employed to predict open reading frames (ORFs) in each assembled contig with a minimum length of 300 bp. A gene catalog without redundancy was created using CD-HIT67 (http://www.bioinformatics.org/cd-hit/, version 4.6.1) with a sequence similarity threshold of 90% and coverage requirement of 90%. Gene abundance was calculated by aligning high-quality reads to non-redundant gene catalogs employing SOAPaligner68 (http://soap.genomics.org.cn/, version 2.21), with a 95% identity threshold.
The analysis of the data was conducted using the online platform provided by Majorbio Cloud Platform (http://www.majorbio.com), which is operated by Shanghai Majorbio Bio−pharm Technology Co., Ltd (China). The representative sequences of the non-redundant gene catalog were aligned to the NR database using DIAMOND69 (http://www.diamondsearch.org/index.php, version 0.8.35) for taxonomic annotations. The COG annotation of the representative sequences was performed using DIAMOND (http://www.diamondsearch.org/index.php, version 0.8.35) by conducting a search against the eggNOG database. The KEGG annotation was performed by employing DIAMOND (http://www.diamondsearch.org/index.php, version 0.8.35) to search against the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/keeg/). The raw metagenomic data have been deposited to the NCBI sequence read archive with accession numbers of SAMN40863842 (30d), SAMN40863843 (60d), and SAMN40863844 (90).
Data availability
Data will be made available on request.
References
Felföldi, T. et al. Polyphasic bacterial community analysis of an aerobic activated sludge removing phenols and thiocyanate from coke plant effluent. Bioresour. Technol. 101, 3406–3414 (2010).
Yu, X., Nishimura, F. & Hidaka, T. Anammox reactor exposure to thiocyanate: Long-term performance and microbial community dynamics. Bioresour. Technol. 317 (2020).
Kanthale, P. et al. Qualitative test for the detection of extraneous thiocyanate in milk. J. Food Sci. Technol. 52, 1698–1704 (2015).
Gould, W. D. et al. A critical review on destruction of thiocyanate in mining effluents. Miner. Eng. 34, 38–47 (2012).
Yu, X.-Z., Lin, Y.-J., Shen, P.-P., Zhang, Q. & Gupta, D. K. Molecular evidences on transport of thiocyanate into rice seedlings and assimilation by 13C and 15N labelling and gene expression analyses. Int. Biodeterior. Biodegrad. 139, 11–17 (2019).
Rahman, S. F. et al. Genome-resolved metagenomics of a bioremediation system for degradation of thiocyanate in mine water containing suspended solid tailings. Microbiologyopen 6, e00446 (2017).
Raper, E., Stephenson, T., Fisher, R., Anderson, D. R. & Soares, A. Characterisation of thiocyanate degradation in a mixed culture activated sludge process treating coke wastewater. Bioresour. Technol. 288, 121524 (2019).
Zaia, D. A. M., de Carvalho, P. C. G., Samulewski, R. B., de Carvalho Pereira, R. & Zaia, C. T. B. V. Unexpected Thiocyanate Adsorption onto Ferrihydrite Under Prebiotic Chemistry Conditions. Orig. Life Evol. Biosphere: J. Int. Soc. Study Orig. Life 50, 57–76 (2020).
Wang, L. et al. Treatment of thiocyanate-containing wastewater: a critical review of thiocyanate destruction in industrial effluents. World J. Microbiol. Biotechnol. 39, 35 (2023).
Phuc, H., Moreau & John, W. Thiocyanate adsorption on ferrihydrite and its fate during ferrihydrite transformation to hematite and goethite. Chemosphere 119, 987–993 (2015).
Sharma, V. K., Yngard, R. A., Cabelli, D. E. & Baum, J. C. Ferrate(VI) and ferrate(V) oxidation of cyanide, thiocyanate, and copper(I) cyanide. Radiat. Phys. Chem. 77, 761–767 (2008).
Wang, X., Liu, L., Lin, W. & Luo, J. Development and characterization of an aerobic bacterial consortium for autotrophic biodegradation of thiocyanate. Chem. Eng. J. 398, 125461 (2020).
Pan, J. et al. Simultaneous removal of thiocyanate and nitrogen from wastewater by autotrophic denitritation process. Bioresour. Technol. 267, 30–37 (2018).
Chen, X., Yang, L., Sun, J., Dai, X. & Ni, B.-J. Modelling of simultaneous nitrogen and thiocyanate removal through coupling thiocyanate-based denitrification with anaerobic ammonium oxidation. Environ. Pollut. 253, 974–980 (2019).
Lee, C., Kim, J., Chang, J. & Hwang, S. Isolation and identification of thiocyanate utilizing chemolithotrophs from gold mine soils. Biodegradation 14, 183–188 (2003).
Boucabeille, C., Bories, A. & Ollivier, P. J. B. L. Degradation of thiocyanate by a bacterial coculture. Biotechnol. Lett. 16, 425–430 (1994).
Malmir, N., Fard, N. A., Aminzadeh, S., Moghaddassi-Jahromi, Z. & Mekuto, L. An Overview of Emerging Cyanide Bioremediation Methods. Processes 10, 1724 (2022).
Kelly, D. P. & Wood, A. P. Confirmation of Thiobacillus denitrificans as a species of the genus Thiobacillus, in the beta-subclass of the Proteobacteria, with strain NCIMB 9548 as the type strain. Int. J. Syst. Evol. Microbiol. 50, 547–550 (2000).
Kataoka, S. et al. Functional expression of thiocyanate hydrolase is promoted by its activator protein, P15K. FEBS Lett. 580, 4667–4672 (2006).
Li, L. et al. Degradation pathway and microbial mechanism of high-concentration thiocyanate in gold mine tailings wastewater. RSC Adv. 10, 25679 (2020).
Stratford, J., Dias, A. E. X. O. & Knowles, C. J. The utilization of thiocyanate as a nitrogen source by a heterotrophic bacterium: the degradative pathway involves formation of ammonia and tetrathionate. Microbiology 140, 2657–2662 (1994).
Pan, J., Wei, C., Fu, B., Ma, J. & Preis, S. Simultaneous nitrite and ammonium production in an autotrophic partial denitrification and ammonification of wastewaters containing thiocyanate. Bioresour. Technol. 252, 20–27 (2018).
Sorokin, D. Y., Tourova, T. P., Antipov, A. N., Muyzer, G. & Kuenen, J. G. Anaerobic growth of the haloalkaliphilic denitrifying sulfur-oxidizing bacterium Thialkalivibrio thiocyanodenitnficans sp nov with thiocyanate. Microbiol.-Sgm 150, 2435–2442 (2004).
Wang, T. et al. Efficient nitrogen removal in separate coupled-system of anammox and sulfur autotrophic denitrification with a nitrification side-branch under substrate fluctuation. Sci. Total Environ. 696, 133929 (2019).
Qian, J. et al. A feasibility study on biological nitrogen removal (BNR) via integrated thiosulfate-driven denitratation with anammox. Chemosphere 208, 793–799 (2018).
Li, X., Yuan, Y., Huang, Y. & Bi, Z. Simultaneous removal of ammonia and nitrate by coupled S0-driven autotrophic denitrification and Anammox process in fluorine-containing semiconductor wastewater. Sci. Total Environ. 661, 235–242 (2019).
Broman, E. et al. Low temperature, autotrophic microbial denitrification using thiosulfate or thiocyanate as electron donor. Biodegradation 28, 287–301 (2017).
Strous, M., Heijnen, J. J., Kuenen, J. G. & Jetten, M. S. M. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl. Microbiol. Biotechnol. 50, 589–596 (1998).
Kuypers, M. M. M. et al. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422, 608–611 (2003).
Kartal, B. et al. Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 30, 39–49 (2007).
Narita, Y. et al. Enrichment and physiological characterization of an anaerobic ammonium-oxidizing bacterium ‘Candidatus Brocadia sapporoensis’. Syst. Appl. Microbiol. 40, 448–457 (2017).
Strous, M. et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794 (2006).
van Niftrik, L. & Jetten, M. S. M. Anaerobic Ammonium-Oxidizing Bacteria: Unique Microorganisms with Exceptional Properties. Microbiol. Mol. Biol. Rev. 76, 585–596 (2012).
Huang, K., Li, Q., Sun, H., Zhang, X. X. & Ye, L. Metagenomic analysis revealed the sulfur- and iron- oxidation capabilities of heterotrophic denitrifying sludge. Ecotoxicology 30, 1399–1407 (2021).
Ting, A. S. Y. et al. Bacterial and eukaryotic microbial communities in urban water systems profiled via Illumina MiSeq platform. 3 Biotech 11, 40 (2021).
Podosokorskaya, O. A. et al. Characterization of Melioribacter roseus gen. nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environ. Microbiol. 15, 1759–1771 (2013).
Xing, W. et al. Identification of the autotrophic denitrifying community in nitrate removal reactors by DNA-stable isotope probing. Bioresour. Technol. 229, 134–142 (2017).
Di Capua, F., Ahoranta, S. H., Papirio, S., Lens, P. N. L. & Esposito, G. Impacts of sulfur source and temperature on sulfur-driven denitrification by pure and mixed cultures of Thiobacillus. Process Biochem 51, 1576–1584 (2016).
Kelly, D. P. & Wood, A. P. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50, 511–516 (2000).
Kantor, S.R. et al. Genome-resolved meta-omics ties microbial dynamics to process performance in biotechnology for thiocyanate degradation. Environ. Sci. Technol. 51, 2944–2953 (2017).
Du, R. et al. Performance and microbial community analysis of a novel DEAMOX based on partial-denitrification and anammox treating ammonia and nitrate wastewaters. Water Res 108, 46–56 (2017).
Oshiki, M., Fukushima, T., Kawano, S., Kasahara, Y. & Nakagawa, J. Thiocyanate Degradation by a Highly Enriched Culture of the Neutrophilic Halophile Thiohalobacter sp. Strain FOKN1 from Activated Sludge and Genomic Insights into Thiocyanate Metabolism. Microbes Environ. 34, 402–411 (2019).
Wang, C., Liu, Y., Wang, C., Xing, B. & Zhu, L. Biochar facilitates rapid restoration of methanogenesis by enhancing direct interspecies electron transfer after high organic loading shock. Bioresour. Technol. 320, 124360 (2021).
Zhang, G. et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 6, 1336–1344 (2012).
Hao, W. et al. Organic carbon coupling with sulfur reducer boosts sulfur based denitrification by Thiobacillus denitrificans. Sci. Total Environ. 748, 142445 (2020).
Tejerizo, G. T. et al. Genome sequence of Methanobacterium congolense strain Buetzberg, a hydrogenotrophic, methanogenic archaeon, isolated from a mesophilic industrial-scale biogas plant utilizing bio-waste. J. Biotechnol. 247, 1–5 (2017).
Zhang, J. et al. Adaptation to salinity: Response of biogas production and microbial communities in anaerobic digestion of kitchen waste to salinity stress. J. Biosci. Bioeng. 130, 173–178 (2020).
Angenent, L. T., Sung, S. W. & Raskin, L. Methanogenic population dynamics during startup of a full-scale anaerobic sequencing batch reactor treating swine waste. Water Res 36, 4648–4654 (2002).
Zhou, L. et al. Transcriptomic and Physiological Insights into the Robustness of Long Filamentous Cells of Methanosaeta harundinacea, Prevalent in Upflow Anaerobic Sludge Blanket Granules. Appl. Environ. Microbiol. 81, 831–839 (2015).
Rotaru, A.-E. et al. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ. Sci. 7, 408–415 (2014).
Kartal, B. et al. Molecular mechanism of anaerobic ammonium oxidation. Nature 479, 127–130 (2011).
Newsholme, P. et al. Glutamine and glutamate as vital metabolites. Braz. J. Med. Biol. Res. 36, 153–163 (2003).
Reinartz, M., Tschäpe, J., Brüser, T., Trüper, H. G. & Dahl, C. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch. Microbiol. 170, 59–68 (1998).
Dahl, C., Franz, B., Hensen, D., Kesselheim, A. & Zigann, R. Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159, 2626–2638 (2013).
Kantor, R. S. et al. Bioreactor microbial ecosystems for thiocyanate and cyanide degradation unravelled with genome-resolved metagenomics. Environ. Microbiol. 17, 4929–4941 (2015).
Ogawa, T. et al. Carbonyl sulfide hydrolase from thiobacillus thioparus strain thi115 is one of the β-carbonic anhydrase family enzymes. J. Am. Chem. Soc. 135, 3818–3825 (2013).
APHA. Standard Methods for the Examination of Water and Sewage. Washington, D. C. (1998).
Tsushima, I., Kindaichi, T. & Okabe, S. Quantification of anaerobic ammonium-oxidizing bacteria in enrichment cultures by real-time PCR. Water Res 41, 785–794 (2007).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Huson, D. H., Mitra, S., Ruscheweyh, H.-J., Weber, N. & Schuster, S. C. Integrative analysis of environmental sequences using MEGAN4. Genome Res 21, 1552–1560 (2011).
Asnicar, F., Weingart, G., Tickle, T. L., Huttenhower, C. & Segata, N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. Peerj 3, e1029 (2015).
Douglas, G. M. et al. PICRUSt2: An improved and customizable approach for metagenome inference. bioRxiv, 672295 (2020).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890 (2018).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 11, 119 (2010).
Noguchi, H., Park, J. & Takagi, T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34, 5623–5630 (2006).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Li, R., Li, Y., Kristiansen, K. & Wang, J. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Meth. 12, 59–60 (2015).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (22076100, 52250410337, 52311540153), Taishan Scholar Youth Expert Program of Shandong Province (tsqn201909005), and Instrument Improvement Funds of Shandong University Public Technology Platform (ts20220106).
Author information
Authors and Affiliations
Contributions
X.C analyzed data and wrote the manuscript. F.D. and X.Y carried out the experiments. Y.Y.X and Z.B.W assisted with the data analysis. S.-Q.N modified the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41545_2024_402_MOESM1_ESM.docx
Uncovering pathway and mechanism of simultaneous thiocyanate detoxicity and nitrate removal through anammox and denitrification
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, X., Duan, F., Yu, X. et al. Uncovering pathway and mechanism of simultaneous thiocyanate detoxicity and nitrate removal through anammox and denitrification. npj Clean Water 7, 109 (2024). https://doi.org/10.1038/s41545-024-00402-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41545-024-00402-w