Abstract
High levels of nitrates and nitrites not only threaten aquatic ecosystems and drinking water safety but also impair the biodegradation efficiency of industrial wastewater. In this study, micro-nano-MoS2-1013 (0.04 g/L) enhanced denitrification by 56.9% and 29.6% in steel pickling and meat processing wastewaters, respectively, and improved chemical oxygen demand (COD) removal by 136.7% in refinery cooling wastewater under continuous-flow conditions using a 3.5 L upflow anaerobic sludge blanket (UASB) reactor. The catalytic effect of micro-nano-MoS2 on denitrification was achieved by stimulating an increase in the abundance of denitrification genes and the transcript levels of narL (93.82 times), narG (16.34 times), and nirK (12.27 times) within the bacterial cells, which led to an increase in the expression levels of denitrifying enzymes. These findings have significant implications for the design and optimization of biodegradation processes and bio-denitrification systems, particularly for the treatment of high-concentration nitrate wastewater.

Similar content being viewed by others
Introduction
With the progression of industrialization, the load, difficulty, and standard requirements for wastewater treatment have rapidly increased, particularly for previously overlooked pollution sources, such as water bodies containing nitrates from factories or agricultural fertilizers. However, nitrates and their secondary metabolites present in water bodies pose a threat to the natural environment and a serious risk to human food safety1,2. The biological denitrification process faces certain complications when applied in the treatment of various industrial wastewaters. For instance, it underperforms in the case of pickling effluents from stainless steel production that contain a high nitrate concentration3. Moreover, it fails to efficiently remove nitrogen in refinery wastewater, despite >93% chemical oxygen demand (COD) removal4, and struggles with high organic and nitrite loads in meat processing wastewater5. These issues underscore the necessity for more robust and efficient denitrification strategies tailored to suit complex industrial effluents. The severity and urgency of this issue is underscored by the slow biodegradation of these pollutants in wastewater treatment plants, which is primarily hindered by the rate-limiting denitrification step6,7,8. Therefore, enhancing denitrification has emerged as a critical area of interest in water treatment9.
In an innovative approach to expedite denitrification, new reactors have been designed to augment biomass, such as the upflow anaerobic sludge bed, moving bed biofilm reactor, and sequencing batch reactor10,11. Certain substances, termed redox mediators12,13, such as anthraquinone, porphyrin, and riboflavin, can also enhance the denitrification process by accelerating electron transfer14,15,16,17. Electroactive bacteria, such as Geobacter sulfurreducens, have also shown potential for accelerating electron transfer, thereby enhancing the denitrification rate by 51%18. In recent years, increasing attention has been directed towards micro- and nano-scale materials for biological nitrogen removal. For instance, reduced graphene oxide has been applied in wastewater treatment to accelerate biological processes, including shortening the start-up time of anammox systems19, improving the total nitrogen removal rate20,21. Graphene- and iron-based nanomaterials significantly enhance anammox activity and promote bacterial growth22. Similarly, micro- and nano-hydroxyapatite have been reported to accelerate aerobic granular sludge formation, reducing the granulation period to 15 and 20 days, respectively23. Although the use of micro- and nano-scale materials has emerged as a promising strategy to enhance denitrification, their practical deployment is hindered by a trade-off between activity and safety. Microscale materials exhibit limited catalytic performance, whereas nanoscale materials—though highly active—are associated with increased toxicity and environmental risks24. However, the underlying biotic-abiotic synergistic mechanism is poorly understood, highlighting the need for a new approach that could directly enhance denitrifying bacteria without causing potential secondary pollution25.
Molybdenum disulfide (MoS2) is valued for its superior photoelectric properties in applications such as photocatalysis and chemical sensing. It also significantly promotes efficacious denitrification in simulated waters due to properties such as efficient electron transfer, structural stability, and minimal environmental impact26,27. Despite this potential, the practical applications of MoS2—especially micro-nano-MoS2—in real industrial wastewater treatment and continuous-flow reactor systems remain largely unexplored. Furthermore, from an industrial application perspective, optimization parameters such as particle size and dosage require systematic evaluation to ensure economic feasibility and effective operational integration.
In this study, we synthesized micro-nano-MoS2 of varying particle sizes and applied them as a bio-denitrification accelerator in industrial wastewater treatment. Their effectiveness in enhancing denitrification under continuous-flow conditions was evaluated using three types of real industrial wastewater. In addition, high-throughput sequencing, shotgun metagenomics, and quantitative PCR (q-PCR) were employed in synthetic wastewater systems to monitor microbial community dynamics, gene abundance, and the transcriptional responses of denitrification-related genes, thereby elucidating microbial stress response mechanisms. Overall, our study provides novel theoretical insights into the biotic–abiotic interactions involved in MoS2-enhanced denitrification and demonstrates its practical potential for utilization in the treatment of industrial wastewater, particularly in challenging scenarios.
Results
The preparation and characteristics of micro-nano-MoS2
A comprehensive characterization of the micro-nano-MoS2 samples was performed using various analytical techniques. Laser granulometry analysis revealed that the particle size distribution of the sample obtained after 5 days of ball milling (micro-nano-MoS2-1013) was centered around a peak of 938.8 nm (Fig. 1a). In contrast, the samples milled for 10 and 15 days exhibited significantly finer particle sizes, with the dominant peaks observed at 584.6 nm (micro-nano-MoS2-1055) and 283.4 nm (micro-nano-MoS2-1236), respectively. However, as each sample contained a small fraction of particles with diameters exceeding 1 µm, the term “micro-nano-MoS2” is deemed most appropriate.
a illustrates the particle size distributions, with different colors representing three independent measurements; b shows the XRD patterns; c presents the FT-IR spectra; d depicts the Mo XPS spectra; e, f provide representative SEM and TEM images, respectively; and g displays the CV curves of the samples.
The X-ray diffraction (XRD) analysis confirmed that all three materials consist of MoS2 particles (Fig. 1b). The Fourier transform-infrared spectroscopy (FT-IR) spectra of the samples exhibited nearly identical characteristic peak positions (Fig. 1c). In the X-ray photoelectron spectroscopy (XPS) analysis, peaks near 229 eV and 232 eV were attributed to Mo atoms in the +4-oxidation state within MoS2 (Fig. 1d). Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) images revealed layered MoS2 structures, and the observed particle size order was consistent with the hydrodynamic analysis (Fig. 1e, f). Cyclic voltammetry (CV) was used to evaluate the electrochemical performance of the materials. Notably, micro-nano-MoS2–1013 exhibited the best performance, with a reduction peak at –0.56 V and an oxidation peak at 1.38 V (Fig. 1g). These results suggest that micro-nano-MoS2–1013, with its superior redox characteristics, is a promising candidate for advanced electrochemical applications. This result is similar to the CV curve of MoS2 reported in the literature28.
The evaluation of micro-nano-MoS2 catalytic activity
Nitrate adsorption onto the micro-nano-MoS2 was tested, and the results indicated that micro-nano-MoS2 did not adsorb nitrate or catalyze its reduction under abiotic conditions. The catalytic performance of the three micro-nano-MoS2 samples (0.05 mg/L) with different particle sizes for denitrification was investigated. All the materials accelerated denitrification (Fig. 2a). The maximum removal rates of inorganic nitrogen (nitrate + nitrite) per unit time were 38.1, 63.5, 47.8, and 60.9 mg/L−¹ h−¹ for the blank and the three MoS2 materials (Fig. 2b). These findings indicate that micro-nano-MoS2-1013 exhibited the highest catalytic activity for denitrification. Therefore, subsequent dose-response experiments were conducted using micro-nano-MoS2-1013.
a shows the NO3−-N concentration changes with different micro-nano-MoS₂ sizes. b presents maximum degradation rate of inorganic nitrogen with different micro-nano-MoS₂ sizes. Effect of the different doses of micro-nano-MoS2-1013 on denitrification in terms of: c NO3−-N concentration; d accumulation of NO2−-N during denitrification; e removal rate of nitrate during denitrification; f removal rate of inorganic nitrogen during denitrification; g maximum degradation rate of nitrate; and h maximum degradation rate of inorganic nitrogen. The inorganic nitrogen concentration was calculated as the sum of the nitrate and nitrite concentrations. Initial conditions of the denitrification experiment: 125 mg/L NO3−-N; 37 °C. The value for each sample is provided as an average with a standard deviation (n = 3).
The denitrification acceleration varied with different doses of micro-nano-MoS2-1013. The experimental groups were labeled M0, M1, M2, M3, M4, M5, and M6, which correspond to the addition of micro-nano-MoS2-1013 to the sludge at 0, 0.04, 0.2, 0.4, 0.8, 1.2, and 2 g/L, respectively.
Figure 2c illustrates the changes in nitrate concentration over time for each group. The nitrate concentrations in M1, M2, M3, M4, M5, and M6 were lower than those in M0 after 2 h. At 6 h, nitrate degradation in M3, M4, M5, and M6 was almost complete. At 8 h, denitrification still occurred in M0, M1, and M2.
As shown in Fig. 2d, the nitrite concentration initially increases and then decreases over the course of the experiment. The maximum accumulation of nitrite was 21.2, 31.6, 12.6, 8.0, 6.6, 6.14, and 8.1 mg/L in M0, M1, M2, M3, M4, M5, and M6, respectively. When the dose of micro-nano-MoS2-1013 exceeded 0.04 g/L, the maximum nitrite accumulation was reduced by 40–68% compared to that in the M0 group.
The nitrate removal rate was calculated based on the differential concentration of nitrate during the first 6 h of batch experiments (Fig. 2e). The nitrate concentration in the groups with micro-nano-MoS2-1013 decreased faster than that in the control group (M0). Compared to M0, the improvement rate of nitrate removal at 6 h was 73.9% in M1. These features were also reflected in the rate of inorganic nitrogen removal, which was calculated based on nitrite degradation during the first 6 h (Fig. 2f). Compared with M0, the rate of nitrate removal in M5 was 136.6%.
To further explore the kinetic patterns of these rate alterations, the temporal variation curves of nitrate and nitrite degradation rates were calculated. The degradation rate of nitrate was the fastest between 4 and 6 h in all experimental groups, except for M1 (Fig. S1). The degradation rate of nitrite was the fastest between 4 and 6 h in all experimental groups (Fig. S2). At 2 h, the nitrate degradation rate peaked in M0 and M1; it peaked 2 h later in M2, M3, M4, M5, and M6 (Fig. 2g). The maximum nitrate degradation rate in M1 was 61.4 mg L−1 h−1, 116.3% higher than that in M0. The maximum nitrite degradation rate in M3 was 64.0 mg L−1 h−1, 119.0% higher than that in M0 (Fig. 2h).
These observations underscore the ability of micro-nano-MoS2-1013 to rapidly enhance the nitrate and nitrite degradation rates in sludge and maintain these rates until the denitrification process is nearly complete. Compared with our previous study of micro-graphite particles (MGPs), it would appear that adding less micro-nano-MoS2-1013 (0.04 g/L) could help sludge achieve the maximum denitrification rate earlier and increase the maximum denitrification rate more than two times. To achieve the same efficiency, the MGPs concentration must be <0.12 g/L.
Studies of micro-nano-MoS2-accelerated bio-denitrification in continuous reactor systems treating industrial wastewaters
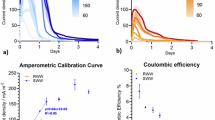
To assess the applicability of micro-nano-MoS2-1013 (0.04 g/L) in complex matrices, preliminary batch experiments were conducted using steel pickling, refinery cooling, and meat processing wastewaters. Despite marked variations in composition and microbial stress, micro-nano-MoS2-1013 consistently enhanced denitrification performance, achieving up to 84.8%, 261.1%, and 171.8% improvements in denitrification efficiency for the three wastewaters, respectively, thereby underscoring its robustness across diverse industrial effluents (Fig. S3).
Guided by the batch results, we further evaluated the catalytic performance of micro-nano-MoS2-1013 (0.15 g) under continuous operation. The UASB reactors were employed to treat the three wastewater types, with operational parameters detailed in Table S2.
During the treatment of steel pickling wastewater, the Micro-nano-MoS2-1013 group exhibited a pronounced acceleration in nitrate degradation, achieving a 36.7% improvement by day 49 (Fig. 3a). Meanwhile, the suppression of nitrite accumulation progressively strengthened, resulting in a 56.9% reduction on day 49. Changes in COD removal were negligible. In the reactor experiments treating refinery cooling wastewater, the nitrate degradation rate in the control group was markedly inhibited by the combined effects of organic nitrogen, toxic compounds, and nitrite accumulation. In contrast, nitrate degradation in the Micro-nano-MoS2-1013 group was significantly accelerated, achieving a 195.3% improvement by day 49 (Fig. 3b). Although severe nitrite accumulation was observed during the initial 20 days, it gradually declined thereafter. Moreover, the COD removal efficiency was enhanced by 1.3-fold by day 49. For the meat processing wastewater, the Micro-nano-MoS2-1013 group exhibited slightly lower and more stable effluent nitrate concentrations (Fig. 3c), accompanied by a marked enhancement in nitrite removal efficiency by 29.6% on day 49, while the trend in COD removal remained largely unchanged. Detailed daily data are presented in Fig. S4.
These results further confirm that Micro-nano-MoS2-1013 retains its catalytic efficacy under continuous flow conditions, reinforcing its potential for practical applications in industrial wastewater denitrification. Importantly, only a single dose of micro-nano-MoS2-1013 (0.15 g) was added at the beginning of the reactor operation, yet its promoting effect on denitrification remained stable throughout the 49-day period.
Molecular biology analysis of sludge under the influence of micro-nano-MoS2
The diversity of the community structure in the sludge samples was sequenced using the Illumina MiSeq platform with 16S rDNA. The dominant genus in the community was Paracoccus with an abundance of 74.17% in the initial sample. After the batch experiment, the proportion of Paracoccus increased by ~3–9% in each group with micro-nano-MoS2 (Figs. 4a, 5b). Micro-nano-MoS2 easily increased the proportion after denitrification. This was consistent with the results of our previous study29.
The genus levels taxonomic classification at a 1 h and b 8 h (excluding general with <1%); the species levels at c 8 h. Samples are named X-Y: X represents the group name; Y represents the sampling time, where 1 h and 8 h correspond to 1 h and 8 h, respectively. The “Initial” samples in (a) and (b) were taken from the flask without any additives that was kept in the same environment as the experimental groups at the beginning and end of the denitrification experiment. d Changes in the abundance of denitrifying genes in sludge exposed to micro-nano-MoS2. The change in the denitrification gene is calculated based on the fold of the sample divided by the initial sample. Red indicates that the fold change is >1; purple indicates that the fold change is <1. Changes in the transcription levels of narL (e), narG (f), nirS (g), and nirK (h) in sludge exposed to micro-nano-MoS2 (n = 6).
To further explore the changes at the bacterial species level, the M0-8 h, M1-8 h, and M5-8 h samples were shotgun sequenced and analyzed using metagenomics. The data for quality control and all sample assembly statistics result are provided in Tables S3 and S4. No obvious differences in alpha diversity were observed among the samples (Table S5). The species-level changes were minimal under the influence of micro-nano-MoS2 (Fig. S5). As shown in Fig. 4c, compared to the initial sample, the increase in Paracoccus abundance is due to the increase in Paracoccus versutus (4.64%) in M0. The increase in Paracoccus abundance in the groups with micro-nano-MoS2 contributed to the increase in P. versutus (2.52%) and Paracoccus denitrificans (0.44%). This suggests that P. versutus and P. denitrificans responded to the presence of micro-nano-MoS2 and increased their abundance. Because these two species are able to degrade nitrate and nitrite30,31, their increase could enhance denitrification.
To further explore the reasons for the changes in community structure and accelerated denitrification, we determined changes in abundance at the gene level by comparing sequences from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. In the immune system, denitrification reduced signaling molecules and interaction pathways by 35.4% and 21.5%, respectively, whereas micro-nano-MoS2 further reduced the immune system by 37.9% (Fig. S6–9). Only M00173 and M00011 decreased slightly after denitrification, regardless of whether it was additional micro-nano-MoS2. There were no obvious differences among samples M1, M2, and M5 (Fig. S10).
Compared to the group without any micro-nano-MoS2, the abundance of several types of denitrifying genes increased in M1, i.e., 3.1%—narG; 4.8%—NRT; 7.3%—narL; 9.2%—nirS; 4.5%—nirK; 8.0%—norB; 8.0%—norC; and 2.8%—nosZ (Fig. 4d). When a larger amount of micro-nano-MoS2 was added, an increase in the abundance of denitrifying genes was observed, i.e., NRT—0.3%; narL—25.8%; nirK—23.2%; norB—0.2%; and nirS—0.3%. Exposure to micro-nano-MoS2increased the abundance of denitrifying enzyme-coding genes. However, a large dose of micro-nano-MoS2 did not cause a further increase in the number of denitrifying genes, except for nirK and narL.
The transcript levels of narL, narG, nirS, and nirK were determined using RT-qPCR (Fig. 4e–h). Within 10 min, the abundance of narL in RNA increased in M1 and M5 by 3.5 and 93.82 times, respectively, compared with that in M0, whereas the transcription level of the narG gene increased in M1 and M5 by 1.57 and 16.34 times, respectively. After 1 h, the abundance of nirK significantly increased in the M1 (2.4 times) and M5 (12.27 times). Under the influence of micro-nano-MoS2, the transcription level of the narL gene was enhanced, and it passed a signal downstream of the effectors (narG, nirS, and nirK) in the presence of nitrate or nitrite. Unlike previously published content, we found that the transcription levels of narG and nirK were upregulated, leading to an enhancement of the denitrification rate27. This pattern likely reflects the stepwise activation of genes in the denitrification pathway. Regulatory genes such as narL respond rapidly to nitrate availability, while downstream genes such as nirK show delayed activation due to intermediate accumulation (e.g., nitrite).
The abundance of pilA (which encodes the type IV pilus assembly protein) increased by 5.9% in the group with micro-nano-MoS2. The abundance of the fliO and fliR genes that encode the flagellar protein and flagellar biosynthesis protein were significantly enhanced by 10.7% and 4.3%, respectively. Collisions between the flagella and micro-nano-MoS2 stimulated the component system, including the narL gene.
The possible pathway of micro-nano-MoS2 accelerating denitrification
According to the above studies, bacteria possess a tactile sense and can perceive mechanical forces32. In the presence of micro-nano-MoS2, surface collisions and interface interactions occur between the material’s active sites and specific proteins on bacterial pili or flagella. The upstream proteins of the two-component system responsible for tactile sensing can trigger downstream signaling cascades (Fig. 5)33. As a downstream component of the two-component system, the narL receptor is influenced by these signals, leading to its upregulation34. narL, a key gene involved in initiating denitrification, positively regulates the transcription of narG and nirK, thereby enhancing their expression35. Under the cascade effect, the increased transcription of denitrification genes promotes the translation of denitrification enzymes, thereby increasing their abundance and accelerating the overall denitrification rate36.
Discussion
This study demonstrates that micro-nano-MoS2 significantly accelerates bio-denitrification by promoting the activity and gene expression of key denitrifying bacteria. In continuous-flow experiments simulating industrial conditions, a single dose of 0.15 g micro-nano-MoS2-1013 maintained strong catalytic effects over 49 days. Specifically, the nitrate degradation efficiency improved by 36.7%, 195.3%, and 171.8% in steel pickling, refinery cooling, and meat processing wastewater, respectively. These improvements were accompanied by effective suppression of nitrite accumulation and stable or improved COD removal, underscoring the material’s robustness and applicability in diverse, complex wastewater environments.
Mechanistically, the interaction between micro-nano-MoS2 and bacterial flagella likely triggers the upregulation of the narL response regulator, which initiates a transcriptional cascade affecting narG, nirS, and nirK. RT-qPCR results confirmed that narL transcript levels rose by 3.5-fold and 93.82-fold in M1 and M5, respectively, within 10 min. This was followed by significant increases in narG and nirK expression, suggesting that MoS2 stimulation rapidly activates the denitrification machinery at the gene expression level. These effects were further supported by metagenomic analyzes, which showed increases in the abundance of denitrifying genes, including narL (25.8%), nirK (23.2%), and norB (8.0%) with increasing MoS2 concentrations.
Compared to other nanomaterials used to enhance denitrification—such as 50 mg/L graphene (67.3% improvement) or 200 mg/L nano-Fe₃O₄ (24.76%)—micro-nano-MoS2 achieved higher enhancement levels at significantly lower doses and with longer-lasting activity37,38. Moreover, no observable leaching of MoS2 occurred, indicating effective retention in the sludge phase and minimizing secondary environmental risks. This allows for the catalyst to be managed together with waste sludge, simplifying downstream handling.
From an application perspective, the material is easily synthesized using scalable ball-milling and annealing methods, requires low dosage, exhibits stable performance over extended durations, and shows strong returns in terms of denitrification efficiency. These features suggest that micro-nano-MoS2-1013 is a cost-effective and operationally viable catalyst for industrial nitrogen removal.
This work forms part of an ongoing research program. Subsequent stages will focus on expanding the reactor scale and further diversifying the range of real industrial wastewater types under investigation. These efforts aim to strengthen the practical foundation for deploying MoS2-assisted denitrification in full-scale treatment facilities.
Methods
Experimental reagents and sludge
All chemical reagents were procured from Aladdin, Ltd. (Shanghai, China). All reagents were of analytical grade and were used without further purification. The sludge was inoculated with activated sludge collected from the Shijiazhuang Qiaoxi Wastewater Treatment Plant. Sludge was cultivated as described in our previous study29.
Preparation of micro-nano-MoS2
In this study, dry ball milling was employed to synthesize micro-nano-MoS2, and three distinct particle size fractions were obtained by controlling the milling duration. Specifically, 500 g of high-purity MoS2 powder was loaded into a stainless-steel ball mill with 5 kg of 10-mm diameter and 2 kg of 20-mm diameter stainless steel balls. Milling was performed at 400 rpm for 5, 10, and 15 days. After 5 days of milling, the MoS2 predominantly consisted of coarser, micrometer-scale particles; 50 g of these particles were collected and designated as micro-nano-MoS2-1013. Extending the milling time to 10 days resulted in a significant reduction in particle size; 50 g of the resultant product was collected and labeled as micro-nano-MoS2-1055. Finally, after 15 days of milling, the MoS2 was further refined, yielding the finest fraction, which was collected and referred to as micro-nano-MoS2-1236.
Characterization of micro-nano-MoS2
The size distribution of the synthesized micro-nano-MoS2 samples was measured using a Mastersizer (LS-POP-6, OMEC, Zhuhai, China). The crystal structures were determined using XRD using a Rigaku Smart Lab9 KW system (Tokyo, Japan). The surface morphology was examined using field-emission SEM (Hitachi S-4800, Tokyo, Japan), and the internal structure was analyzed using TEM (JEOL JEM-2100, Tokyo, Japan). The surface chemical composition and functional groups were characterized using XPS (Thermo Fisher K-Alpha, MA, USA) and FT-IR (PerkinElmer SP3, MA, USA), respectively. Additionally, CV measurements were performed using an electrochemical workstation (Chenhua CH1660E, Shanghai, China) to evaluate the electrochemical properties of the samples.
Assessment of nitrate adsorption and abiotic reduction by micro-nano-MoS2
The adsorption and abiotic reduction of nitrate onto micro-nano-MoS2 was tested to determine whether the micro-nano-MoS2 interfered with the nitrate concentration analysis results. After being exposed to 50 mg/L micro-nano-MoS2, synthetic wastewater with an initial concentration of 100 mg/L NO3−-N did not show a decrease in nitrate concentration after 8 h.
Enhancement of denitrification by micro-nano-MoS2 in batch experiments
Synthetic wastewater was prepared by mixing sodium succinate and nitrate with cultivated sludge, following our previous method [23]. After mixing, the final concentrations of CODCr and NO₃−-N were 750 mg/L and 125 mg/L, respectively. A total volume of 250 mL of the mixture was transferred into conical flasks and incubated at 37 °C under anaerobic conditions. Two separate batch experiments were conducted to evaluate the effects of micro-nano-MoS2: one to compare different particle sizes, and another to assess different dosages (0.04–2 g/L). Flasks without micro-nano-MoS2 served as the control group. To calculate the denitrification rate, all groups were sampled every 2 h until most of the denitrification in the experimental group was complete. Changes in the concentrations of pollutants (NO3−-N and NO2−-N) and the pH of the synthetic wastewater were determined immediately. All experiments were performed in triplicate. Nitrate, nitrite, and pH measurements were performed as previously reported29.
Enhancement of denitrification by micro-nano-MoS2 with industrial wastewater in batch experiments
Three types of real industrial wastewater samples were obtained from full-scale chemical production plants (steel pickling wastewater, refinery cooling wastewater, and meat processing plants wastewater) to assess the denitrification catalytic capacity of micro-nano-MoS2-1013. The nitrate and nitrite concentrations (mg/L as NO₃−-N/NO2⁻-N) were 68.3/0.6, 334.8/11.6, and 81.4/184 in steel pickling, refinery cooling, and meat processing plant wastewater, respectively (other component details are available in Table S1). The microbial inoculum was prepared by centrifugation (8000 × g, 10 min) of 250 mL fermentation broth, followed by resuspension in 10 mL sterile phosphate-buffered saline (0.1 M, pH 7.4).
Batch experiments were conducted with 250 mL Erlenmeyer flasks containing 240 mL of wastewater amended with 10 mL of concentrated inoculum. The experimental groups received 0.2 g L−¹ micro-nano-MoS2-1013 catalyst, whereas control groups maintained equivalent systems without catalyst addition. To address insufficient BOD5, glucose was supplemented as a carbon source by adding 0.281 g/L (equivalent to 300 mg/L COD) in the steel pickling wastewater experiment and 1.406 g/L (equivalent to 1500 mg/L COD) in the refinery cooling wastewater experiment. Experiments were performed in triplicate with identical operational parameters.
Enhancement of denitrification by micro-nano-MoS2 with industrial wastewater in reactor systems
In the reactor experiments, three types of industrial wastewater—steel pickling wastewater, refinery cooling wastewater, and meat processing plant wastewater—were used to evaluate the ability of micro-nano-MoS2 to enhance denitrification. To address the issue of insufficient BOD₅, glucose was supplemented as an additional carbon source: 0.281 g/L (equivalent to 300 mg/L COD) was added in the steel pickling wastewater experiments, and 1.406 g/L (equivalent to 1500 mg/L COD) was added in the refinery cooling wastewater experiments. A 3.5 L UASB reactor was employed, with the upflow velocity detailed in Table S2. Influent and effluent concentrations of nitrate, nitrite, and COD were monitored daily throughout the experiments. All experiments were conducted in triplicate.
16S rRNA gene-based analysis
The sludge was sampled at the beginning and end of the experiment. All samples were sequenced using the Illumina-MiSeq platform. High-throughput sequencing was performed as described in our previous study29.
Metagenomic analysis
DNA was extracted from the samples at the beginning and the end of the experiments for metagenomic sequencing using an EZNA® Mag-Bind Soil DNA Kit, following the manufacturer’s instructions. The metagenomics data were sequenced on a HiSeq sequencer (Illumina) at Sangon Biotech® to generate paired-end reads. The raw DNA sequences can be found on the National Microbiology Data Center (NMDC) website under the NMDC number NMDC10017878.
For functional annotation, representative sequences from the non-redundant gene catalog were aligned to the KEGG database. The relative abundance of functional genes was determined as the proportion of matching reads assigned to a category in the total effective reads, as previously described39.
q-PCR analysis
Total RNA was extracted from the samples at the end of the experiment using the Sangon Biotech® Bacteria Total RNA Isolation Kit (Shanghai, China). Reverse transcription of RNA was performed using the Sangon Biotech® MightyScript Plus First Strand cDNA Synthesis Master Mix (gDNA digester). The abundance of four target genes was quantified, including three denitrification genes (narG, nirS, and nirK) and 16S rDNA using the SYBR-Green q-PCR method with 2 × SGExcel Fast SYBR Mixture (with ROX, Sangon Biotech). The PCR procedure for 16S rDNA included an initial denaturation step at 94 °C for 30 s, followed by 40 cycles of amplification (94 °C for 30 s, 57 °C for 30 s, 72 °C for 2 min), and a final extension step at 72 °C for 10 min. The final holding temperature for preservation was 4 °C. The primers used were as follows:
16S: ATGGCTGTCGTCAGCT, ACGGGGCGGTGTGTAC.
NarG: GACAAACTTCGCAGCGG, TCACCCAGGACGCTGTTC.
NirS: CCTA(C/T)TGGCCGCC(A/G)CA(A/G)T, GCCGCCGTC(A/G)TG(A/C/G)AGGAA.
NirK: GG(A/C)ATGGT(G/T)CC(C/G)TGGCA, GCCTCGATCAG(A/G)TT(A/G)TGG.
All processes used in the experiments were performed according to the manufacturer’s instructions and amplified in six replicates.
Statistical analysis
Data are expressed as the mean values of the standard deviation of three biological replicates. Statistical significance of the changes was determined using Tukey’s multiple range test (Excel). The significant correlation between the dose and acceleration was determined using Pearson’s correlation coefficient. Statistical significance was set at p < 0.05.
Data availability
Raw DNA sequences are available at the National Microbiology Data Center (NMDC) under accession number NMDC10017878.
References
Bougarne, L., Ben Abbou, M., El Haji, M. & Bouka, H. Consequences of surface water eutrophication: Remedy and environmental interest. Mater. Today Proc. 13, 654–662 (2019).
Wang, Z. et al. Spatial association of surface water quality and human cancer in China. NPJ Clean Water 6, 53 (2023).
Zeng, B., Jiang, Y., Pan, Z., Shen, L. & Lin, H. Feasibility and optimization of a novel upflow denitrification reactor using denitrifying granular sludge for nitric acid pickling wastewater treatment. Bioresour. Technol. 384, 129271 (2023).
Kenari, H. R., Sarrafzadeh, M. H. & Tavakoli, O. An investigation on the nitrogen content of a petroleum refinery wastewater and its removal by biological treatment. Iran. J. Environ. Health Sci. Eng. 7, 391–394 (2010).
Aziz, A. et al. Biological wastewater treatment (anaerobic-aerobic) technologies for safe discharge of treated slaughterhouse and meat processing wastewater. Sci. Total Environ. 686, 681–708 (2019).
Skiba, U. & Hydrology, C. Alcaligenes faecalis, Pseudomonas stutzeri, Paracoccus denitrificans, 866–871 (2008).
Yanai, Y., Toyota, K. & Okazaki, M. Effects of charcoal addition on N2O emissions from soil resulting from rewetting air-dried soil in short-term laboratory experiments: original article. Soil Sci. Plant Nutr. 53, 181–188 (2007).
Ren, W., Ren, G., Teng, Y., Li, Z. & Li, L. Time-dependent effect of graphene on the structure, abundance, and function of the soil bacterial community. J. Hazard. Mater. 297, 286–294 (2015).
McCarty, P. L. What is the best biological process for nitrogen removal: when and why?. Environ. Sci. Technol. 52, 3835–3841 (2018).
Dai, C. et al. Promoting the granulation process of aerobic granular sludge in an integrated moving bed biofilm-membrane bioreactor under a continuous-flowing mode. Sci. Total Environ. 703, 135482 (2020).
Daija, L. et al. The influence of lower temperature, influent fluctuations and long retention time on the performance of an upflow mode laboratory-scale septic tank. Desalin. Water Treat. 57, 18679–18687 (2016).
Lovley, D. R., Coates, J. D., Blunt-Harris, E. L., Phillips, E. J. P. & Woodward, J. C. Humic substances as electron acceptors for microbial respiration. Nature 382, 445–448 (1996).
Peng, C. et al. Enhanced biofilm formation and denitrification in biofilters for advanced nitrogen removal by rhamnolipid addition. Bioresour. Technol. 287, 121387 (2019).
Aranda-Tamaura, C. et al. Effects of different quinoid redox mediators on the removal of sulphide and nitrate via denitrification. Chemosphere 69, 1722–1727 (2007).
Martins, L. R., Baêta, B. E. L., Gurgel, L. V. A., de Aquino, S. F. & Gil, L. F. Application of cellulose-immobilized riboflavin as a redox mediator for anaerobic degradation of a model azo dye Remazol Golden Yellow RNL. Ind. Crops Prod. 65, 454–462 (2015).
Xi, Z. et al. Study the catalyzing mechanism of dissolved redox mediators on bio-denitrification by metabolic inhibitors. Bioresour. Technol. 140, 22–27 (2013).
Xie, Z. et al. Biocatalysis mechanisms and characterization of a novel denitrification process with porphyrin compounds based on the electron transfer chain. Bioresour. Technol. 265, 548–553 (2018).
Zhou, L. et al. Subminimal inhibitory concentration (sub-MIC) of antibiotic induces electroactive biofilm formation in bioelectrochemical systems. Water Res. 125, 280–287 (2017).
Yin, X., Qiao, S., Zhou, J. & Tang, X. Fast start-up of the anammox process with addition of reduced graphene oxides. Chem. Eng. J. 283, 160–166 (2016).
Tomaszewski, M., Cema, G. & Ziembińska-Buczyńska, A. Short-term effects of reduced graphene oxide on the anammox biomass activity at low temperatures. Sci. Total Environ. 646, 206–211 (2019).
Yin, X., Qiao, S., Yu, C., Tian, T. & Zhou, J. Effects of reduced graphene oxide on the activities of anammox biomass and key enzymes. Chem. Eng. J. 276, 106–112 (2015).
Wang, S., Zhang, K., Miao, Y. & Wang, Z. The enhancement of anammox by graphene-based and iron-based nanomaterials in performance and mechanisms. NPJ Clean Water 7, 100 (2024).
Shi, W. et al. Deciphering the role of micro/nano-hydroxyapatite in aerobic granular sludge system: effects on treatment performance and enhancement mechanism. J. Environ. Manag. 366, 121850 (2024).
Bae, M. et al. Nanotoxicity of 2d molybdenum disulfide, mos2, nanosheets on beneficial soil bacteria, bacillus cereus and pseudomonas aeruginosa. Nanomaterials 11, 1453 (2021).
Bian, J. et al. Synergy of cyano groups and cobalt single atoms in graphitic carbon nitride for enhanced bio-denitrification. Water Res. 218, 118465 (2022).
Oviroh, P. O., Jen, T.-C., Ren, J. & van Duin, A. Towards the realisation of high permi-selective MoS2 membrane for water desalination. NPJ Clean Water 6, 14 (2023).
Zhang, Y., Sun, H., Lu, C., Li, H. & Guo, J. Role of molybdenum compounds in enhancing denitrification: Structure-activity relationship and the regulatory mechanisms. Chemosphere 367, 143433 (2024).
Hu, R., Huang, Z., Wang, B., Qiao, H. & Qi, X. Electrochemical exfoliation of molybdenum disulfide nanosheets for high-performance supercapacitors. J. Mater. Sci. Mater. Electron. 32, 7237–724 (2021).
Li, J. et al. Micro-graphite particles accelerate denitrification in biological treatment systems. Bioresour. Technol. 308, 122935 (2020).
Olaya-Abril, A. et al. Effect of pH on the denitrification proteome of the soil bacterium Paracoccus denitrificans PD1222. Sci. Rep. 11, 17276 (2021).
Shi, Z., Zhang, Y., Zhou, J., Chen, M. & Wang, X. Biological removal of nitrate and ammonium under aerobic atmosphere by Paracoccus versutus LYM. Bioresour. Technol. 148, 144–148 (2013).
Hug, I., Deshpande, S., Sprecher, K. S., Pfohl, T. & Jenal, U. Second messenger-mediated tactile response by a bacterial rotary motor. Science 358, 531–534 (2017).
Chen, Y. T., Chang, H. Y., Lu, C. L. & Peng, H. L. Evolutionary analysis of the two-component systems in Pseudomonas aeruginosa PAO1. J. Mol. Evol. 59, 725–737 (2004).
Ruanto, P. et al. Activation by NarL at the Escherichia coli ogt promoter. Biochem. J. 477, 2807–2820 (2020).
Yang, Y., Wang, J., Xiu, Z. & Alvarez, P. J. J. Impacts of silver nanoparticles on cellular and transcriptional activity of nitrogen-cycling bacteria. Environ. Toxicol. Chem. 32, 1488–1494 (2013).
Torres, C. I. et al. Selecting anode-respiring bacteria based on anode potential: phylogenetic, electrochemical, and microscopic characterization. Environ. Sci. Technol. 43, 9519–9524 (2009).
Jiang, M., Feng, L., Zheng, X. & Chen, Y. Bio-denitrification performance enhanced by graphene-facilitated iron acquisition. Water Res. 180, 115916 (2020).
Chen, Y. et al. Role of Nano-Fe3O4 for enhancing nitrate removal in microbial electrolytic cells: characterizations and microbial patterns of cathodic biofilm. Chemosphere 339, 139643 (2023).
Huang, W. et al. Metagenomic analysis reveals enhanced nutrient removal from low C/N municipal wastewater in a pilot-scale modified AAO system coupling electrolysis. Water Res. 173, 115530 (2020).
Acknowledgments
The authors are grateful for financial assistance received from the National Basic Research Program of China (No. 2012CB723501), the National Natural Science Foundation of China (No. 21978067) and the Hebei Natural Science Foundation (No.12966737D). We thank Yuqing Ma for assistance in creating the figures. We thank Sangon Biotech (www.sangon.com) for assistance in DNA sequencing.
Author information
Authors and Affiliations
Contributions
Z.P. performed formal analysis, investigation, prepared visualizations, and wrote the original draft. J.S. and P.L. contributed to the investigation. J.L. was responsible for conceptualization, methodology, resources, project administration, and writing—review and editing. C.S., P.K., T.M., D.H., and X.L. contributed to writing—review and editing. X.T. and X.Z. provided supervision. J.L. and X.L. provided resources. S.L. contributed to resources, writing—review and editing, and funding acquisition. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peng, Z., Sun, J., Li, P. et al. Boosting industrial bio-denitrification through gene activation: the micro-nano-MoS2 enhanced bioprocess. npj Clean Water 8, 63 (2025). https://doi.org/10.1038/s41545-025-00494-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41545-025-00494-y