Abstract
Current protocols for the differentiation of human pluripotent stem cells (hPSCs) into chondrocytes do not allow for the expansion of intermediate progenitors so as to prospectively assess their chondrogenic potential. Here we report a protocol that leverages PRRX1–tdTomato reporter hPSCs for the selective induction of expandable and ontogenetically defined PRRX1+ limb-bud-like mesenchymal cells under defined xeno-free conditions, and the prospective assessment of the cells’ chondrogenic potential via the cell-surface markers CD90, CD140B and CD82. The cells, which proliferated stably and exhibited the potential to undergo chondrogenic differentiation, formed hyaline cartilaginous-like tissue commensurate to their PRRX1-expression levels. Moreover, we show that limb-bud-like mesenchymal cells derived from patient-derived induced hPSCs can be used to identify therapeutic candidates for type II collagenopathy and we developed a method to generate uniformly sized hyaline cartilaginous-like particles by plating the cells on culture dishes coated with spots of a zwitterionic polymer. PRRX1+ limb-bud-like mesenchymal cells could facilitate the mass production of chondrocytes and cartilaginous tissues for applications in drug screening and tissue engineering.
Similar content being viewed by others
Main
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), are widely employed in developmental biology and regenerative medicine because of their indefinite proliferative capacity and pluripotency. Limb articular hyaline cartilage is composed of type II collagen-producing chondrocytes and defects in, or degeneration of, these chondrocytes due to trauma or necrosis results in the loss of joint function, such as osteoarthritis. Difficulties in the regeneration of cartilaginous tissues have focused research efforts on employing hPSCs to establish chondrogenic induction protocols. Skeletal tissues, including the cartilage and bone, originate from ontogenetically defined mesodermal tissues of vertebrates as follows: paraxial mesoderm (PM) for the axial skeleton (for example, vertebrae), neural crest for the cranial skeleton (for example, cranial bones) and lateral plate mesoderm (LPM) for the appendicular skeleton (for example, limbs)1,2. Several methods for the induction of PM- or neural crest-derived chondrocytes have been developed3,4,5,6,7; however, none of these methods include quality tests to determine whether intermediate progenitor cells exhibit bona fide phenotypic features of chondrogenic progenitors in vivo. These technical difficulties represent critical disadvantages that hinder the efficient and stable production of cells and experimental reproducibility. Moreover, the lack of methods to expand and cryopreserve intermediate progenitors of chondrocytes impedes the mass production of chondrocytes/cartilaginous tissues of sufficient quality and causes insufficient processability for tissue engineering of cartilaginous tissue for application in regenerative medicine.
During limb development, the LPM generates the limb-bud mesenchyme (LBM), in which chondrogenesis is initiated in a timely manner via a stringently regulated signal transduction network8,9,10,11,12. PRRX1 serves as a marker for the LBM, and Prrx1-deficient mice as well as PRRX1 mutations in humans were shown to exhibit lethal phenotypes, characterized by craniofacial defects and limb shortening13,14,15,16. In mouse LBM, PRRX1+ cells are the progenitors of SOX9+ cells, which serve as the precursor of osteo-chondroprogenitor cells17. Genetic lineage tracing studies using Prrx1–Cre transgenic mice clearly demonstrated that PRRX1+ cells are the source of limb chondrocytes, which are essential for endochondral ossification18,19. Although mouse LBM cells isolated from the developing mouse limb bud possess high chondrogenic capacity17,20, human LBM cells have not been assessed for chondrogenesis because of a lack of methods to induce their differentiation from hPSCs.
In this study we describe the development of a protocol to generate ontogenetically defined, expandable PRRX1+ LBM-like (ExpLBM) cells from hPSCs under defined xeno-free conditions. ExpLBM cells stably proliferate and exhibit a high potential to generate hyaline cartilaginous tissues in vitro. We further identify CD90highCD140BhighCD82low cell populations as ExpLBM cells with high chondrogenic differentiation potential. ExpLBM cells can be applied to disease modelling of patients with type II collagenopathy (COL2pathy). Finally, we describe an ExpLBM-based high-throughput screening system and a simple protocol that facilitates the production of numerous uniform-sized hyaline cartilaginous particles using a specific charged polymer.
Results
Induction of ontogenetically defined PRRX1+ cells from hPSCs under defined xeno-free conditions
To visualize PRRX1+ cells at each step of differentiation, we designed a targeting vector harbouring IRES–tdTomato–PGK–Neo cassettes for the bicistronic expression of PRRX1 and tdTomato (Fig. 1a). We electroporated 414C2 iPSCs with the pCR8-3′PRRX1 HA–IRES–tdTomato–PGK–Neo targeting vector and PX459-PRRX1 guide RNA (gRNA) genome-editing vector. The electroporated cells were selected with G418, followed by single-cell cloning to isolate PRRX13′tdTomato-reporter iPSCs (Fig. 1b). Single colonies were expanded and genomic DNA was extracted to identify the correct insertion of the knock-in reporter cassettes. A clone harbouring a homozygous knock-in of the reporter cassette was identified through PCR analysis using primers that recognise sequences flanking the homology arms (HA; Fig. 1c). This PRRX13′tdTomato-reporter clone expressed the pluripotency markers SSEA4, NANOG, OCT4 and SOX2 (Supplementary Fig. 1a,b), and was therefore selected for further experiments.
a, Targeting cassette of the hPRRX13′tdTomato knock-in allele. PAM sequence (CCT) is described in red. b, Generation of the PRRX13′tdTomato-reporter hiPSC line. The targeting and gRNA–Cas9 expression vector were electroporated into the hiPSC line 414C2. After selection with G418, single colonies were selected, expanded and screened for the presence of the knock-in reporter cassette. c, Agarose gel of a genotyping PCR of the genomic DNA extracted from the parental 414C2 hiPSCs and PRRX13′tdTomato-reporter iPSCs. Targeting cassette sequences were detected using forward and reverse primers that recognise sequences outside the HAs. WT, wild-type allele; KI, knock-in allele. d, Overview of the stepwise induction and differentiation of human LBM mimicking embryonic development. e, Immunocytochemical analysis of HAND1 (LPM marker) and CDX2 (PM marker) expression by PRRX13′tdTomato-reporter-derived LPM (day 2; left) and PRRX13′tdTomato-reporter-derived PM (day 2; right). The nuclei were counterstained with DAPI. Representative images are shown. f, Expression of tdTomato in PRRX13′tdTomato-reporter-derived LPM cells (day 2) treated with 16 combinations of chemicals and cytokines for 2 d, as described in Supplementary Table 1. WNT signal on, CHIR (GSK3β inhibitor, 3 μM); WNT signal off, C59 (PORCN inhibitor, 1 μM); BMP signal on, BMP4 (30 ng ml−1); BMP signal off, LDN-193189 (ALK2/3 inhibitor, 0.5 μM); TGF-β signal on, TGF-β1 (10 ng ml−1); TGF-β signal off, A-83-01 (ALK4/5/7 inhibitor, 1 μM); HH signal on, SAG21K (SMO agonist, 5 nM) and HH signal off, vismodegib (SMO inhibitor, 0.15 μM). Cells that were stimulated with CHIR99021 (WNT signal on) expressed tdTomato. g, Comparison of PRRX1 expression after LPM cells were treated with each combination of chemicals and cytokines. Total RNA was extracted on days 0, 1, 2 and 4, and the expression levels of marker genes were compared using RT–qPCR. h, Comparison of the levels of the LPM-marker genes HAND1, FOXF1 and ISL1 following the treatment of LPM cells with the various combinations of chemicals and cytokines. Total RNA was extracted at days 0, 1, 2 and 4, and the levels of marker genes were compared using RT–qPCR. g,h, All expression values are normalized to those of ACTB mRNA (n = 3 biologically independent experiments). i, Flow cytometry analysis of tdTomato expression in PRRX13′tdTomato-reporter cells at pluripotency (day 0), LPM (day 2) and the day-4 (combination 8) state. j, PRRX1 expression in PRRX13′tdTomato-reporter cells at pluripotency (day 0), LPM (day 2) and day 4 (combination 8). The levels of PRRX1 at each time point were compared using immunoblotting (n = 3 biologically independent experiments). k, Immunostaining of PRRX1 in the PRRX13′tdTomato-reporter cells at pluripotency (day 0) and the PRRX1+ state. All cells co-expressed PRRX1 and tdTomato. g,h, ND, not detected; *P < 0.05; ***P < 0.001. Statistical significance was determined using a one-way ANOVA with correction for multiple testing using the Bonferroni method.
Mesoderm development begins with the differentiation of pluripotent epiblast cells in the primitive streak; these cells then segregate into PM and LPM21,22. To establish an induction protocol using ontogenetically defined LBM cells, we first performed stepwise induction of hPSCs to generate middle primitive streak and LPM23. We then determined the optimal conditions to induce the differentiation of LBM cells into PRRX1+ cells using the PRRX13′tdTomato-reporter (Fig. 1d). Immunocytochemical analysis showed that the two-step induction from the pluripotent state (day 0) produced HAND1+ (LPM marker) and CDX2− (PM marker) cells on day 2 (Fig. 1e) through the transitional primitive-streak state (Supplementary Fig. 1c).
Next, PRRX13′tdTomato-reporter-derived LPM cells (day 2) were treated with 16 different combinations of chemicals and recombinant proteins for 2 d to modify four signalling pathways: WNT, BMP, TGF-β and hedgehog (HH; Supplementary Table 1). WNT activation (combinations 1–8) was required to induce tdTomato expression (Fig. 1f) and PRRX1 messenger RNA expression (Fig. 1g) in LPM cells. During limb-bud development, WNT signalling promotes the proliferation, and inhibits the apoptosis, of LBM cells24,25. The TGF-β, BMP and HH signalling pathways mediate the differentiation or apoptosis of LBM cells20,26,27, suggesting that activation of WNT signalling and inhibition of the other three pathways induce the differentiation of PRRX1+ cells from LPM cells. Furthermore, the expression levels of LPM-marker genes (HAND1 (ref. 28), FOXF1 (ref. 29) and ISL1 (ref.30)) were significantly downregulated only when BMP, TGF-β and HH signalling were inhibited (Fig. 1h). We therefore selected combination 8 (WNT activation; BMP, TGF-β and HH inhibition) for further experiments. Flow cytometry revealed that most LPM-derived cells expressed tdTomato (96.6 ± 1.8%; Fig. 1i) and that the levels of PRRX1 protein were increased (Fig. 1j), corroborating the transition from LPM to the PRRX1+ state by combination 8. Colocalization of PRRX1 and tdTomato indicated that the reporter system correctly visualized PRRX1+ cells (Fig. 1k). Other hPSCs, including hiPSCs (414C2 and 1383D2) and hESCs (SEES4, SEES5, SEES6 and SEES7), demonstrated similar PRRX1 immunostaining on days 0 and 4 (Extended Data Fig. 1) and PRRX1, HAND1, FOXF1 and ISL1 expression levels—determined by reverse transcription quantitative PCR (RT–qPCR)—at each differentiation stage (Supplementary Fig. 2).
Expansion and characterization of PRRX1+ LBM-like cells under defined xeno-free conditions
The development of a protocol to expand progenitor cells is required for the mass production of differentiated cells with high processability into the desired sizes and shapes suitable for application in tissue engineering. To assess the stable proliferation of PRRX1+ cells, we tested three types of culture media (mediums 1–3), which are described in Fig. 2a. Only medium 3 (hereinafter referred to as ExpLBM medium) supported the proliferation of LBM-like cells after the first passage while maintaining PRRX1–tdTomato expression (Fig. 2a). These expandable LBM-like cells (called ExpLBM cells) did not exhibit altered morphology during serial passage (Fig. 2b) and stably expressed PRRX1 (Fig. 2c). Furthermore, ExpLBM cells retained both the expression levels and positive rate of PRRX1–tdTomato during serial passage (Fig. 2d). We found that 2 × 105 LBM-like cells, harvested on day 4, yielded 1.2 × 1015 ExpLBM cells after ten passages (doubling time, 2.3 ± 0.2 d). The generation of PRRX1+ ExpLBM cells with similar doubling times was reproduced using other hPSCs (doubling times: SEES4, 3.0 ± 0.2 d; SEES5, 4.3 ± 0.8 d; SEES6, 2.7 ± 0.2 d and SEES7, 2.8 ± 0.2 d; Fig. 2e,f and Supplementary Fig. 3a). We also confirmed the normal karyotype of ExpLBM cells (Supplementary Fig. 3b). Interestingly, ExpLBM cells could be stored in liquid nitrogen without a decrease in the levels of PRRX1–tdTomato expression (Supplementary Fig. 4a).
a, Serial passage of LBM-like cells stably expressing PRRX1. After collecting PRRX13′tdTomato-reporter line-derived LBM-like cells from the culture dish, the cells were seeded and cultured in medium 1 (LBM medium, combination 8), medium 2 (LBM medium containing 20 ng ml−1 epidermal growth factor (EGF) and 20 ng ml−1 fibroblast growth factor (FGF)) or medium 3 (medium 2 excluding LDN-193189 and vismodegib; ExpLBM medium). Cell morphology and tdTomato fluorescence were observed 1 or 5 d after seeding. LBM-like cells were passaged while maintaining tdTomato expression using medium 3 (ExpLBM medium). b–d, Cell morphology (b), PRRX1 immunostaining (c) and tdTomato fluorescence intensity (d) of ExpLBM cells derived from the PRRX13′tdTomato-reporter line at each passage number when the cells were maintained in ExpLBM medium. The cells (2 × 105) were passaged in 6 cm culture dishes every week. e,f, Growth curve (e) and PRRX1 immunostaining (f) of ExpLBM cells derived from the reporter and SEES4–7 (e), and SEES4 and -5 hPSC lines. g,h, RNA transcriptome analysis of pluripotent stem cells (day 0), LPM (day 2), LBM-like (day 4) and the ExpLBM (PN = 4 or 5) state (PRRX13′tdTomato-reporter-derived samples; n = 2 biologically independent experiments). Principal component (PC) analysis (g) and heatmap of gene expression levels of each marker (h).
To determine the cellular identity of ExpLBM cells, we performed RNA-sequencing (RNA-seq) analysis of sequential samples at each step of the generation of ExpLBM cells from PRRX13′tdTomato-reporter hiPSCs. Principal component analysis revealed distinct differential gene expression among pluripotent (day 0), LPM (day 2), LBM-like (day 4) and ExpLBM cells (Fig. 2g). The expression levels of the pluripotent (UTF1, GDF3, NANOG, GRB7, DNMT3B, TDGF1, DPPA4, SOX2 and PODXL), LPM (FOXF1, ISL1, HAND1, TBX20 and GATA4) and LBM (PRRX1, LMO2, TSHZ2, DUSP7, MSX1, HAND2, PITX1, PRDM1, GLI3, ZFHX4, LHX2, LMX1B, FIGF, HOXA9, HOXA10, HOXC5, HOXC9, HOXC10, HOXC11, HOXD9, HOXD10 and HOXD11) marker genes clearly revealed the sequential induction of genes that were representative of each differentiation stage under the established culture conditions. We found that the expansion procedures increased the expression levels of the LBM marker genes, indicating that culturing with ExpLBM medium confers an LBM-like phenotype on cells at differentiation day 4 (Fig. 2h). These results suggest that our differentiation and expansion method can be used to generate ontogenetically defined PRRX1+ LBM-like cells from hPSCs.
Chondrogenic capacity of ExpLBM cells
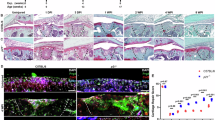
In mice, SOX9+ LBM cells in the developing limb bud are progenitors of chondrocytes17. Flow cytometry analysis revealed that ExpLBM cells derived from each hPSC line were exclusively SOX9+ (Fig. 3a). To determine the chondrogenic capacity of ExpLBM cells, we employed two-dimensional (adhesive culture) or three-dimensional (floating culture) chondrogenic induction protocols (2DCI and 3DCI, respectively) under defined xeno-free conditions. During limb-bud development towards endochondral ossification, the WNT and FGF signalling pathways are required for the condensation of LBM cells, which initiate timely chondrogenesis10,24,25. Therefore, we included the following steps in the chondrogenic induction protocol applied to ExpLBM cells: condensation (step 1), chondrogenic differentiation (step 2) and maturation (step 3). When chondrogenic induction was performed using 2DCI (Fig. 3b), PRRX13′tdTomato-reporter-derived ExpLBM cells produced several nodules that were stained by Alcian blue. These nodules were obtained only when BMP4, TGF-β1, GDF5 and FGF2 were included in the step 2 medium (Fig. 3c). Analysis by RT–qPCR revealed that the expression levels of the cartilaginous tissue marker genes SOX5, SOX6, SOX9, COL2A1 and ACAN were significantly upregulated (Fig. 3d).
a, Flow cytometry analysis of SOX9 expression in ExpLBM cells derived from various hPSCs. b, Two-dimensional chondrogenic induction (2DCI) protocol for ExpLBM cells. c, Alcian blue staining after the 2DCI-based differentiation of ExpLBM cells. After culturing PRRX13′tdTomato-reporter line-derived ExpLBM cells with step 1 medium for 6 d, the cells were treated with step 2 medium for 6 d. The cells were then fixed and stained with Alcian blue (n = 3 biologically independent experiments). d, Analysis of chondrocyte marker genes during the 2DCI protocol. Total RNA was extracted at the indicated time points and the gene expression levels were determined by RT–qPCR analysis. All values were normalized to those of ACTB mRNA (n = 3 biologically independent experiments). ND, not detected. c,d, **P < 0.01; ***P < 0.001; NS, not significant. Statistical significance was determined using a two-sided unpaired Student’s t-test (c) or one-way ANOVA with correction for multiple testing using the Bonferroni method (d). e, Three-dimensional chondrogenic induction (3DCI) protocol for ExpLBM cells. f, Images of hPSCs-derived particles using the 3DCI protocol. g, Histological analysis of particles generated using the 3DCI protocol. Particles were generated from ExpLBM cells derived from each hPSC, fixed and then stained with haematoxylin–eosin (HE), Alcian blue or Safranin O, or subjected to immunohistological analysis using antibodies against SOX9, COL2, COL1 or COLX. f,g, Data for day 42 of step 3. h, Subcutaneous transplantation of cartilaginous particles in NOD-SCID mice. Cartilaginous particles (414C2-derived; step 3 treatment, 21 d) were subcutaneously transplanted into NOD-SCID mice and their engraftment was assessed by HE, Alcian blue or Safranin O staining at 8 weeks after transplantation. c,f–h, Representative images are shown.
In the 3DCI protocol (Fig. 3e), each hPSC-derived ExpLBM cell reproducibly generated white particles at day 42 of step 3, 42 d (Fig. 3f). Histological analysis showed that the cells were embedded in numerous regions of the extracellular matrix (ECM), which was stained intensely and homogeneously with Alcian blue and Safranin O (Fig. 3g, second and third columns). Immunohistological analysis revealed that all cells embedded in the ECM expressed SOX9; type II collagen (COL2) was detected in the ECM but type I collagen (COL1) and type X collagen (COLX) were not (Fig. 3g, four columns on the right). Analysis by RT–qPCR revealed that the expression levels of chondrocyte (SOX5, SOX6, SOX9, COL2A1 and ACAN) and hypertrophic (COL10A1, IHH and MMP13) marker genes were upregulated in the 3DCI protocol (Supplementary Fig. 5a). The 3DCI protocol considerably upregulated the expression of not only pan-chondrocyte marker genes (SOX5, SOX6, SOX9, COL2A1, COL9A1, COL9A2, COL9A3, COL11A1, COL11A2 and ACAN), but also articular cartilage (GDF5, PRG4, CILP, CILP2 and CLU) and hypertrophic (COL10A1, IHH and MMP13) marker genes, compared with the 2DCI protocol, as determined by RNA-seq analysis (Supplementary Fig. 5b,c and Supplementary Table 2). Furthermore, the capacity of ExpLBM cells to form cartilaginous tissue under these conditions was maintained after cryopreservation (Supplementary Fig. 4b). When ExpLBM cells were subjected to the 3DCI protocol without step 1, the ECM within the particles was lightly stained with Alcian blue and Safranin O (Supplementary Fig. 6a). In contrast to ExpLBM cells, LBM-like cells on day 4 of differentiation formed particles containing a heterogeneous cell population in which the ECM was not stained by Safranin O (Supplementary Fig. 6b).
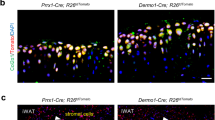
To test the graft survival of ExpLBM-derived particles, we performed the following transplantation assays. ExpLBM-derived particles (step 3, 21 d) that were subcutaneously transplanted into NOD-SCID mice maintained hyaline cartilage-like structures without any teratoma development at 8 weeks after transplantation (Fig. 3h and Supplementary Figs. 7a–i and 8a). We also transplanted PRRX13′tdTomato-reporter-derived cartilaginous particles (step 3, 21 d) to a 1 mm hole defect in the articular cartilage of SCID rats and tested their engraftment. As shown in Extended Data Fig. 2 and Supplementary Fig. 8a, human antigen (human VIMENTIN)-positive hyaline cartilage-like tissues were observed at 4 weeks after transplantation without any teratoma development. Importantly, the colony-forming assay revealed that single cells isolated from cartilaginous particles (step 3, 21 d) could not form any colonies, indicating that the cells in the cartilaginous particles did not possess tumorigenic capacity (Supplementary Fig. 8b). When subcutaneous transplantation was performed in NOD-SCID mice with βTCP, human bone marrow mesenchymal stem cells, but not PRRX13′tdTomato-reporter-derived ExpLBM cells, formed bone-like tissues 4 weeks after transplantation (Extended Data Fig. 3a–c). Thus, our ExpLBM-derived cartilaginous particles can serve as a potential regenerative resource of hyaline cartilage tissues.
Identification of CD antigens to enable the assessment of the chondrogenic potential of ExpLBM cells
Methods to prospectively determine the differentiation capacity of intermediate progenitors have become necessary for stable and sufficient supply as well as for reproducibility in the field of biomedical engineering. We therefore attempted to develop a method to enable the prospective chondrogenic assessment of ExpLBM cells. We found that if ExpLBM cells were allowed to attain confluency, this led to an irreversible decrease in the levels of PRRX1–tdTomato and eventually generated PRRX1–tdTomatolow (or PRRX1low) ExpLBM cells (Fig. 4a). Although the conditions necessary to induce PRRX1low ExpLBM cells from PRRX1high ExpLBM cells have not been defined in detail, we investigated the differences between PRRX1high and PRRX1low ExpLBM cells in their capacity to undergo chondrogenic differentiation. In the 2DCI protocol, PRRX1low ExpLBM cells did not form Alcian blue-positive nodules and the staining intensities of PRRX1low ExpLBM cells were significantly lower than those of PRRX1high ExpLBM cells (Fig. 4b). In the 3DCI protocol, PRRX1low ExpLBM cells did not detectably form hyaline cartilaginous-like tissues, in contrast to PRRX1high ExpLBM cells (Fig. 4c). These findings suggest that the identification of several cell-surface markers with a positive or negative correlation with PRRX1 expression will be useful for predicting the chondrogenic potential of ExpLBM cells.
a, Effect of high-density culture on the stable maintenance of ExpLBM cells derived from the PRRX13′tdTomato-reporter line. ExpLBM cells were cultured until they attained high confluency and then analysed to detect tdTomato expression during serial passage. All cells eventually expressed low levels of tdTomato (PRRX1low ExpLBM cells). b, Levels of Alcian blue staining after 2DCI with PRRX1high and PRRX1low ExpLBM cells (right; n = 3 biologically independent experiments). Representative images are shown (left). c, Histological analysis after 3DCI with PRRX1high (top) and PRRX1low (bottom) ExpLBM cells. d, Volcano plot identifying genes that are differentially expressed between PRRX1high and PRRX1low ExpLBM cells. The dashed vertical and horizontal lines indicate the threshold. The red dots represent genes that are differentially expressed (n = 2 biologically independent experiments). The gene names of each identified surface antigen are highlighted in blue. e, Representative flow cytometric analysis data for CD90 (THY1), CD140B (PDGFRB) and CD82 expressed by PRRX1high and PRRX1low ExpLBM cells. f, Representative flow cytometric data for CD90 (THY1), CD140B (PDGFRB) and CD82 expression of SEES4- and 1383D2-derived ExpLBM cells. g, Histological analysis after 3DCI with SEES4- (left) and 1383D2 (right)-derived ExpLBM cells. h,i, CD90highCD140BhighCD82low and CD90lowCD140BlowCD82high populations that were purified from 414C2-derived ExpLBM cells using a cell sorter were treated with the 2DCI protocol, followed by Alcian blue staining (h) and RT–qPCR analysis (i; n = 3 biologically independent experiments). h, Representative images (left) and the levels of Alcian blue staining (right) are shown. b,h,i, *P < 0.05; **P < 0.01; ***P < 0.001. Statistical significance was determined using a two-sided unpaired Student’s t-test.
To identify candidate cell-surface markers to enable distinction between PRRX1high and PRRX1low ExpLBM cells, we conducted RNA-seq analysis of the global gene expression. A volcano plot analysis was performed to identify genes that were differentially expressed between the PRRX1high and PRRX1low ExpLBM cells (Fig. 4d and Supplementary Table 3). The expression levels of genes characteristically expressed in LBM (PRRX1, DUSP7, FIGF, HAND2, HOXA10, HOXC10, LHX2, LMO2, LMX1B, MSX1, PITX1 and PRDM1) were upregulated in the PRRX1high ExpLBM cells (Fig. 4d). We identified 22 genes encoding cell-surface markers that were upregulated in the PRRX1high ExpLBM cells and 18 that were upregulated in the PRRX1low ExpLBM cells. Flow cytometry analysis revealed that CD90 (THY1) and CD140B (PDGFRB) were expressed in conjunction with PRRX1–tdTomato. In contrast, CD82 expression was inversely correlated with PRRX1–tdTomato expression (Fig. 4e). When ExpLBM cells derived from SEES4 or 1383D2 were stained with CD90, CD140B or CD82, the SEES4-derived ExpLBM cells had staining patterns similar to those of the PRRX1high ExpLBM cells; however, the 1383D2-derived ExpLBM cells showed heterogeneous staining patterns for CD90 and CD140B (Fig. 4f). In the 3DCI protocol, the SEES4-derived ExpLBM cells formed hyaline cartilaginous-like tissues but the 1383D2-derived ExpLBM cells did not (Fig. 4g). In the 2DCI assay, the CD90highCD140BhighCD82low populations sorted from the 414C2-derived ExpLBM cells (Supplementary Fig. 9) demonstrated higher intensities of Alcian blue staining (Fig. 4h) and higher expression levels of chondrocyte marker genes (SOX5, SOX6, SOX9, COL2A1, ACAN, IHH and MMP13) than the CD90lowCD140BlowCD82high populations (Fig. 4i). These results demonstrate that CD90highCD140BhighCD82low cell-surface expression indicates ExpLBM cells with high chondrogenic differentiation potential and therefore, the chondrocyte differentiation potential of ExpLBM cells can be prospectively assessed using these CD antigens.
In vitro disease modelling of COL2pathy using ExpLBM cells and development of the ExpLBM-based high-throughput screening system
In disease modelling, the development of a protocol to produce the desired patient-derived cell types with both reproducibility and mass productivity is critical towards achieving high-throughput drug screening in drug discovery. To evaluate the feasibility of ExpLBM cells for the analysis of genetic diseases, we generated ExpLBM cells from COL2pathy-derived hiPSCs31 and assessed their chondrocyte differentiation potential. Type II collagenopathy arising from mutations in COL2A1 comprises a wide spectrum of highly variable phenotypes32,33. Achondrogenesis type II (ACGII) and hypochondrogenesis (HCG) are lethal and the corresponding patient-derived iPSCs undergo abnormal chondrogenic differentiation31. ExpLBM cells were established from ACGII-1 and HCG-1 hiPSCs (Fig. 5a). Immunocytochemical analysis of the ACGII-1- and HCG-1-derived ExpLBM cells revealed that they were exclusively PRRX1+ (Fig. 5b). ACGII-1- and HCG-1-derived ExpLBM cells showed similar doubling times to 414C2 ExpLBM cells (doubling times: 414C2, 2.9 ± 0.1 d; ACGII-1, 3.9 ± 0.6 d; and HCG-1, 3.6 ± 0.5 d; Fig. 5c).
a, Experimental procedures to establish ExpLBM cells from iPSC lines derived from patients with COL2pathy (ACGII-1 and HCG-1 ExpLBMs). b,c, Immunocytochemical analysis of PRRX1 expression (b) and growth curves (c) of ACGII-1 (b,c), HCG-1 (b,c) and 414C2 (c) ExpLBMs. d, Levels of Alcian blue staining after 2DCI with 414C2, Reporter, SEES4, SEES5, SEES7, ACGII-1 and HCG-1 ExpLBMs. After culturing with step 2 medium for 6 d, the cells were fixed and stained with Alcian blue (left). Then the absorbances at 570 nm were compared (right; n = 3 biologically independent experiments). e, Analysis (RT–qPCR) of the expression of chondrocyte marker genes after 2DCI of each ExpLBM cell type. All values were normalized to those of ACTB mRNA (n = 3 biologically independent experiments). f, Chondrogenic index of each chemical. ACGII-1-derived ExpLBM cells were seeded into 96-well plates and differentiated using the 2DCI protocol. Each chemical was added to the step 2 medium at 5 µM (BML-2865, natural compounds; and BML-2843J, FDA-approved drugs) and the chondrogenic indexes were calculated to compare their chondrogenic activities 6 d after chemical treatment. g, Second screening of the identified chemicals. ACGII-1-derived ExpLBM cells were seeded into 96-well plates and differentiated using the indicated procedure. Each chemical was added to the step 2 medium at 0.1 µM. After culturing with step 2 medium for 6 d, the cells were fixed and stained with Alcian blue (left). Then the absorbances at 570 nm were compared (right; n = 3 independent experiments). b,d,g, Representative images are shown. d,e,g, **P < 0.01; ***P < 0.001; NS, not significant. Statistical significance was determined using a one-way (d,g) or two-way (e) ANOVA with correction for multiple testing using the Bonferroni method.
When chondrogenic differentiation was induced using the 2DCI protocol, ACGII-1- and HCG-1-derived ExpLBM cells formed fewer Alcian blue-stained nodules and the staining intensities were significantly lower than that of other ExpLBM control cells (414C2 and PRRX13′tdTomato-reporter hiPSCs, and SEES4, SEES5 and SEES7 hESCs; Fig. 5d and Extended Data Fig. 4a). Moreover, the expression of chondrocyte marker genes was not upregulated in either cell line using the 2DCI protocol (Fig. 5e).
As shown in Extended Data Fig. 4b,c, ACGII-1- and HCG-1-derived ExpLBM cells could form particle structures but they were sparsely stained with Alcian blue, Safranin O and COL2A1, even if the cells in the particles were exclusively SOX9+. In addition, the ACGII-1-derived particles expressed lower levels of SOX5, SOX6, SOX9 and ACAN than the ExpLBM control-derived particles (Extended Data Fig. 4d). Consistent with previous findings31, electron microscopy revealed a distended endoplasmic reticulum in chondrocytes (Extended Data Fig. 5) in the particles derived from ACGII-1 and HCG-1 ExpLBM cells.
To identify therapeutic candidates for COL2pathy, we established a simplified chemical screening system based on the 2DCI protocol (Supplementary Fig. 10). ACGII-1-derived ExpLBM cells had a lower chondrogenic capacity than healthy ExpLBM cells; therefore, we selected ACGII-1-derived ExpLBM cells to identify chemicals that increase their capacity for chondrocyte differentiation. Chemicals derived from the BML-2865 (502 natural compounds) and BML-2843J (765 FDA-approved drugs) library were used. After culturing ACGII-1-derived ExpLBM cells with step 1 medium, they were treated with step 2 medium containing each chemical at a concentration of 5 µM for 6 d. Next, we defined the chondrogenic index as the nodule area in each well/control well (empty well). We found that several chemicals had a chondrogenic index of more than twofold (Fig. 5f). Five of the chemicals identified in this screening (catanospermine, mevastatin, felodipine, nimodipine and bicalutamide) were confirmed to increase the intensity of Alcian blue staining (Fig. 5g). These results showed that the ExpLBM cells were suitable for use in a high-throughput screening system.
Development of a simple protocol for the mass production of uniform-sized hyaline cartilaginous-like tissues using ExpLBM cells
A protocol to mass-produce uniform-sized cartilaginous particles is valuable for the regeneration of a wide range of human articular cartilage defects or high-throughput drug screening for cartilage disease. Zwitterionic copolymers34,35,36 induce cell self-aggregation following cell adhesion, condensation and delamination, thereby enabling the isolation of defined aggregates bound to polymer spots on the culture surface. We developed a protocol to produce numerous ExpLBM-derived cartilaginous particles of a uniform size using a polymer (Fig. 6a). PRRX13′tdTomato-reporter-ExpLBM cells proliferated and condensed only on polymer-coated spots and were co-expressed with PRRX1–tdTomato (Fig. 6b). Following culture using the 3DCI protocol in the presence of SphereMAX to prevent the particles from fusing, the condensed cells formed numerous uniform-sized particles (Fig. 6c,d). These particles were homogeneously stained with Alcian blue, Safranin O and antibodies against SOX9 and COL2 but not with antibodies against COL1 and COLX (Fig. 6e). We observed successful engraftment of these particles four weeks after transplantation in the articular cartilage defects of SCID rats (Fig. 6f). These results suggest that ExpLBM cells are a promising cell source for the mass production of hyaline cartilaginous-like tissues with a desired size/shape when combined with cell self-aggregation technology.
a,b, Experimental procedure for the mass production of uniform-sized cartilaginous particles. a, Ultralow-attachment dishes were coated with droplets of zwitterionic copolymers at intervals of 0.5 mm using a droplet spotter. Reporter-ExpLBM cells were added to polymer droplet-coated dishes and cultured in ExpLBM medium for 2 d, followed by fluorescence microscopy to observe the expression of tdTomato (b). Following cell self-aggregation with delamination, all cell aggregates were transferred to ultralow attachment culture plates and cultured with 3DCI medium in the presence of 0.04% SphereMAX. c,d, Images of mass-produced ExpLBM-derived cartilaginous particles. d, Magnified views of the mass-produced ExpLBM-derived cartilaginous particles. e, Histological analysis of mass-produced ExpLBM-derived cartilaginous particles. Particle sections were stained with HE, Alcian blue and Safranin O, or with antibodies against SOX9, COL2, COL1 and COLX. f, Engraftment of PRRX13′tdTomato-reporter-derived cartilaginous particles, generated using zwitterionic copolymers, into an articular cartilage defect in SCID rats. The knee joints were fixed at 4 weeks after transplantation and tissue sections were stained with HE, Safranin O and toluidine blue. b–f, Representative images are shown. g, Schematic view of the hierarchical induction of ExpLBM-derived chondrocytes.
Discussion
Our stepwise induction protocol (Fig. 6g) is based on the principle that a desired cell type is generated at lineage bifurcation by activating the signals that induce a given fate and/or repressing the activating signals that induce an alternate fate. Research on vertebrate developmental biology using model organisms has identified signals essential for limb-bud formation from LPM. For example, at LPM bifurcation into LBM or cardiac mesoderm, inhibition of WNT signalling is required for cardiac mesoderm specification in LPM in vivo37. Furthermore, activation of BMP and FGF signalling is required to induce the differentiation of cardiac mesoderm from hPSCs-derived LPM23. Our finding of the induction of PRRX1+ LBM-like cells (day 4) from LPM (day 2) corroborates the requirement for WNT activation and BMP blockade, which inhibits the cardiac mesoderm specification in LPM. In contrast, WNT and FGF signals interact to coordinate the outgrowth of LBM cells in the distal direction during limb development24 and the TGF-β signalling pathway defines the fate of LBM cells in chondrogenesis, leading to endochondral ossification27. Our expansion procedures, in which WNT and FGF signalling were activated and TGF-β signalling was inhibited, recapitulated the developmental processes required for limb-bud outgrowth that precedes chondrogenesis. In fact, the count values obtained from the RNA-seq analysis of 3DCI-treated ExpLBM (step 3, 42 d; Supplementary Fig. 5c) showed that ExpLBM-derived cartilaginous tissues were composed of a mixture of articular and hypertrophic chondrocytes, suggesting that ExpLBM cells can generate both articular cartilage and growth-plate cartilage tissues as well as in vivo LBM cells. Thus, our protocol not only recapitulated the in vivo signalling pathway that drives lineage bifurcation but also provides insights into signalling pathways and chromatin dynamics during human limb development, thereby shedding light on the pathogenesis of congenital skeletal diseases.
Although several studies have reported the induction of LBM-like cells from mouse PSCs38,39 (Supplementary Table 4), we developed and validated a method to induce ontogenetically defined human LBM-like cells with the capacity to form homogeneous hyaline cartilaginous-like tissues from hPSCs. Just as important is the recapitulation of the developmental processes that induce the desired cell types, which is intimately associated with the epigenetic state of the transplanted cells. Induction of the desired terminally differentiated cells without recapitulating developmental processes may lead to unexpected results because epigenetic modifications occur during development or differentiation40. Studies of abnormal epigenetic modifications in various diseases, including cancer41,42,43,44, demonstrated the requirement of an appropriate epigenetic state at each differentiation step of hPSCs. Therefore, our ontogenetically defined ExpLBM cells may serve as a safe resource for generating engraftable chondrocytes/cartilaginous tissues that can be utilized in regenerative medicine.
The ontogenetically defined induction protocol for generating LBM-like cells established in this study is indispensable for the reconstitution of disease pathologies employing hiPSCs from patients with hereditary skeletal diseases. Thus, our system recapitulated the pathological phenotypes of COL2pathy that typically affect the limbs and axial skeletogenesis. Although cell-context dependency in pathogenic processes is the focus of studies of tumorigenesis45,46, a recent study reported cell-context dependency when recapitulating the disease phenotypes using iPSCs derived from a patient with fibrodysplasia ossificans progressiva, which is characterized by endochondral heterotopic ossification in soft tissues7. Therefore, the utility of our induction system is applicable to assess the key features of human limb development and address the causative molecular mechanisms associated with skeletal disease that specifically affect limb skeletogenesis in a cell context-dependent manner.
Several protocols have been developed to induce the differentiation of chondrocytes or cartilaginous tissues from hPSCs via PM5, mesendoderm3,4,6 or neural crest/ectomesenchyme-derived progenitors6 (summarized in Supplementary Table 4); however, they lack the ability to evaluate the differentiation capacity of the intermediate progenitors of chondrocytes. ExpLBM cells can be passaged and cryopreserved without a loss of chondrogenic capacity. In particular, the chondrogenic capacity of ExpLBM cells can be prospectively assessed by analysing the expression of the cell-surface markers CD90, CD140B and CD82. Compared with established hPSC-derived chondrocyte-induction protocols that include a protocol for generating expandable SOX9-expressing CD271+PDGFRα+CD73+ chondrogenic ectomesenchymal cells6, our protocols are unique because they yield expandable and cryopreserved homogeneous chondrocyte progenitors that enable the prospective assessment of the chondrogenic capacity required for forming homogenous hyaline cartilaginous-like tissues. Considering that our procedure induces hPSCs to form terminally differentiated chondrocytes using defined xeno-free culture media without animal serum, our method using ExpLBM cells can yield a stable and sufficient supply of chondrocyte/cartilaginous tissues in vitro with high reproducibility.
In biomedical tissue engineering using cartilage tissues for application in regenerative medicine and high-throughput drug screening, the development of protocols to achieve mass productivity and high processability is critically important3,4,47. Most cell sources in this field are largely based on the utility of human mesenchymal stromal cells, which are expanded on culture dishes following their isolation from human tissues48,49. However, difficulties are caused by quality assessment, which is largely dependent on the donor characteristics and the limitation of mass production of quality-assured human mesenchymal stromal cells with chondrogenic capacity50,51,52. Thus, the ExpLBM cells used in our protocol confer the advantage of enabling expansion through serial passage while maintaining its quality regarding the chondrogenic differentiation capacity through the analysis of cell-surface markers. Furthermore, a single plating of ExpLBM cells on a culture dish coated with many droplets of cell self-aggregation-inducible specific charged polymers form numerous uniform-sized hyaline cartilaginous-like tissues. This supports the use of ExpLBM cells in biomedical tissue engineering. We also showed that COL2pathy patient-derived ExpLBM cells are useful for the identification of therapeutic candidates when combined with our 2DCI protocol (Fig. 5). Our screening protocol includes an easy evaluation method for drug screening, demonstrating that disease-specific ExpLBM cells can be specialized for establishing therapeutic strategies.
We have shown that ontogenetically defined human LBM-like cells with prospective assessment of chondrogenic capacity can be generated from hPSCs. Our protocol may facilitate the study of human skeletal development and hereditary skeletal diseases, and may also contribute to regenerative medicine and the discovery of drugs targeting cartilage-related diseases.
Methods
Cell culture
The hPSCs were cultured and maintained using StemFit (AK02N, Ajinomoto). Before attaining subconfluency, the cells were dissociated with TrypLE Select (Thermo Fisher) in 0.25 mM EDTA and suspended in StemFit containing 10 µM Y27632. The cells (1 × 104) were then suspended in StemFit containing 10 µM Y27632 and 8 µl iMatrix511 (human laminin-511 E8 fragment; Nippi) and added to a 6 cm dish. The culture media were replaced the next day with fresh StemFit without Y27632. The media were changed every 2 d until the next passage. The following hPSC lines were used: 414C2 hiPSCs (provided by the Center for iPS Cell Research and Application, Kyoto University); and 1383D2 hiPSCs and SEES4, SEES5, SEES6 and SEES7 hESCs (donated by RIKEN BRC, Japan). The COL2pathy-derived hiPSCs, including ACGII-1 hiPSCs (heterozygous 1 bp substitution (T > C) located at the acceptor site of exon 41 in COL2A1) and HCG-1 hiPSCs (heterozygous 1348G > C (Gly450Arg) mutation in exon 21 in COL2A1), were provided by N. Tsumaki (Center for iPS Cell Research and Application, Kyoto University). Detailed information on the hiPSCs is provided in Supplementary Table 5. Human bone marrow mesenchymal stem cells and HeLa cells were purchased from LONZA (lot no. 0000494678) and ATCC (ATCC CCL-2), respectively.
Generation of a PRRX13′tdTomato-reporter cell line
The targeting strategy for the knock-in of the IRES–tdTomato–PGK–Neo cassette at the PRRX1 3′ untranslated region (UTR) to achieve bicistronic expression of PRRX1 and tdTomato is shown in Fig. 1a. The PRRX1 3′ homology region (PRRX1 3′HA) was synthesised using PCR using a KOD FX NEO kit (Takara). Genomic DNA extracted from 414C2 hiPSCs, which served as a template, was ligated with pCR8/GW/TOPO using TA cloning (Thermo Fisher). To construct the targeting vector (pCR8-3′PRRX1 HA–IRES–tdTomato–PGK–Neo), IRES–tdTomato–PGK–Neo fragments were inserted into the pCR8-3′PRRX1 HA vector using an In-Fusion HD cloning kit (Takara). Guide RNAs were designed to target the PAM sequence located 200 bp downstream from the stop codon of PRRX1 (CCTCTGGGATACCACCACCACTT; the PAM sequence is indicated in bold font). The gRNA oligos (Supplementary Table 6) were designed, phosphorylated, annealed and subcloned into the PX459 vector (Addgene, cat. no. 62988) harbouring a Cas9 expression cassette (PX459-PRRX1 gRNA). To generate the PRRX13′tdTomato-reporter line, 1 × 106 414C2 iPSCs were electroporated with the pCR8-3′PRRX1 HA–IRES–tdTomato–PGK–Neo vector (7.5 μg) and PX459-PRRX1 gRNA (2.5 μg). Selection with G418 (Life Technologies) was maintained for 10 d until stable colonies appeared. Colonies with a diameter >500 μm were selected using a P200 pipette tip with the aid of a microscope and used to inoculate larger volumes of media. To verify precise integration of the IRES–tdTomato–PGK–Neo cassette into the PRRX1 3′ UTR locus, the 5′ or 3′ ends between PRRX1 and IRES–tdTomato–PGK–Neo cassette were amplified for nucleotide sequence analysis. The PCR primers used are listed in Supplementary Table 6.
Differentiation of LBM-like cells
Human pluripotent stem cells (3 × 104) were suspended in 1 ml of StemFit (Ajinomoto) containing 10 µM Y27632 (Wako) and added to a 3.5 cm culture dish containing 4 µl iMatrix511. The culture medium was replaced the next day with fresh StemFit without Y27632. After culturing for 2 d, the cells were washed with PBS and differentiation was induced by changing the culture medium at each time point. Chemically defined CDM2 medium23 was used as the basal culture medium to which cytokines or other chemicals were added to prepare each differentiation medium. PRRX13′tdTomato-reporter-derived LPM was treated for 2 d with 16 combinations of chemicals that were identified as affecting signal transduction as well as cytokines (Supplementary Table 1; WNT signal on, CHIR99021 (GSK3β inhibitor, 3 μM); WNT signal off, C59 (PORCN inhibitor, 1 μM); BMP signal on, BMP4 (30 ng ml−1); BMP signal off, LDN-193189 (ALK2/3 inhibitor, 0.5 μM); TGF-β signal on, TGF-β1 (10 ng ml−1); TGF-β signal off, A-83-01 (ALK4/5/7 inhibitor, 1 μM); HH signal on, SAG21K (SMO agonist, 5 nM) and HH signal off, vismodegib (SMO inhibitor, 0.15 μM)). The composition of the CDM2 basal medium was as follows: 50% IMDM (+GlutaMAX; Gibco) + 50% F12 (+GlutaMAX; Gibco) + 1 mg ml−1 polyvinyl alcohol (Sigma-Aldrich) + 1% (vol/vol) chemically defined lipid concentrate (Gibco) + 450 μM monothioglycerol (Sigma-Aldrich) + 7 μg ml−1 insulin (Sigma-Aldrich) + 15 μg ml−1 transferrin (Sigma-Aldrich) + 1% (vol/vol) penicillin–streptomycin (Gibco). The composition of cytokines or other chemicals in the media was as follows: mid-primitive streak, CDM2 basal medium + 30 ng ml−1 activin A + 40 ng ml−1 BMP4 + 6 µM CHIR99021 + 100 nM PIK90 + 20 ng ml−1 FGF2 + 10 µM Y27632; LPM, CDM2 medium + 1 µM A-83-01 + 30 ng ml−1 BMP4 + 1 µM C59 + 10 µM Y27632; LBM-like, CDM2 basal medium +1 µM A-83-01 + 0.5 µM LDN-193189 + 3 µM CHIR99021 + 150 nM vismodegib + 10 µM Y27632; anterior primitive streak, CDM2 basal medium + 30 ng ml−1 activin A + 4 µM CHIR99021 + 20 ng ml−1 FGF2 + 100 nM PIK90 + 10 µM Y27632; and PM, CDM2 medium + 1 µM A-83-01 + 3 µM CHIR99021 + 0.25 µM LDN-193189 + 20 ng ml−1 FGF2 + 10 µM Y27632. The details of the human recombinant proteins and small molecule agonists or inhibitors are listed in Supplementary Table 7.
Immunocytochemistry
Cells cultured on dishes were fixed with 4% paraformaldehyde (PFA) for 30 min at room temperature and then incubated with blocking solution (3% normal goat serum and 0.1% Triton X-100 in PBS) for 1 h at room temperature. The primary or secondary antibodies were diluted 200- or 500-fold with blocking solution and added to the cell cultures for 1 h at room temperature. After incubation, the nuclei were stained with 0.1 μg ml−1 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher). The samples were observed using a BZ-X710 fluorescence microscope (Keyence). The antibodies used are listed in Supplementary Table 8.
RT–qPCR
Total RNA was extracted using an RNeasy kit (Qiagen) and complementary DNA was synthesised using M-MLV reverse transcriptase (Thermo Fisher) and oligo-dT primers (Sigma-Aldrich). The cDNAs were then used as templates for qPCR analysis with gene-specific primers. The qPCR was performed using an AriaMX real-time PCR system (Agilent). The cycle parameters were as follows: denaturation at 95 °C for 30 s, annealing at 62 °C for 30 s and elongation at 72 °C for 30 s. The expression level of each gene was calculated using the \(2^{{\Delta\Delta}{C_t}}\) method. The primer sequences are described in Supplementary Table 5.
Flow cytometry and cell sorting
Following dissociation with Accutase (Thermo Fisher), 1 × 105 cells were suspended in 100 µl of 2% FBS in PBS and incubated with primary antibody (diluted 200×) for 1 h on ice. The cells were then washed and suspended in 100 µl of 2% FBS in PBS containing secondary antibody (diluted 1,000×) for 30 min on ice. SOX9 was stained using a transcription factor buffer set (BD) following the manufacturer’s instructions. Fluorescence was detected using a CytoFLEX S flow cytometer (Beckman Coulter) and data were analysed using the FlowJo software. For cell sorting, the cells were stained with FITC–CD90, BB700–CD140B and PE-Cy7–CD82, followed by the purification of CD90highCD140BhighCD82low or CD90lowCD140BlowCD82high populations using a BD FACSAria III cell sorter (BD Biosciences). The antibodies used are listed in Supplementary Table 8.
Immunoblotting
Proteins were extracted with lysis buffer (0.1 M Tris, pH 6.7 and 4% SDS) and quantified using a BCA protein assay kit (Thermo Fisher) by measuring the absorbance at 450 nm using a Multiskan Sky microplate spectrophotometer (Thermo Fisher). The proteins (10 µg) were separated using SDS–PAGE and electrophoretically transferred to 0.45 µm PVDF membranes (Millipore), which were blocked with 5% (wt/vol) skim milk and 0.02% (vol/vol) Tween 20 in PBS and incubated overnight with primary antibodies (diluted 1:2,000) at 4 °C. After incubation with horseradish peroxidase-conjugated antibodies (diluted 1:5,000; GE Healthcare), the membranes were reacted with ECL Prime (GE Healthcare) to detect signals using an Amersham Imager 600. The antibodies used are listed in Supplementary Table 8.
Serial passage of ExpLBM cells
Accutase (Thermo Fisher) was used to dissociate LBM-like cells (day 4), and 2 × 105 cells were suspended in ExpLBM medium (CDM2 basal medium + 1 µM A-83-01 + 3 µM CHIR99021 + 20 ng ml−1 FGF2 + 20 ng ml−1 epidermal growth factor + 10 µM Y27632) containing 16 µl iMatrix511 and then added to a 6 cm culture dish. The culture media were replaced with fresh ExpLBM medium every 2 d. Before attaining subconfluency, the cells were passaged as described above.
RNA-seq analysis
Total RNA was extracted using an RNeasy kit (Qiagen), and sequencing libraries were prepared using a KAPA RNA HyperPrep kit with RiboErase (HMR; Kapa Biosystems) and a SeqCap adapter kit (Set A or Set B; Roche) following the manufacturers’ instructions. The sequencing libraries were transferred to a GENEWIZ and loaded onto a HiSeq 2500 system (Illumina) for sequencing. All sequence reads were extracted in FASTQ format using the CASAVA 1.8.4 pipeline. Trimmomatic (version 0.36) was used to remove the adaptors and filter raw reads that were smaller than 36 bases as well as leading and trailing bases with quality below 20. The filtered reads were mapped to hg19 using the HISAT2 software (version 2.1.0). Raw counts for each gene were based on sense-strand data obtained using the featureCounts software from the Subread package. RUVSeq (release 3.10) was used for further normalization to account for sample variations. Differentially expressed genes were identified using DESeq2 analysis with a threshold of an adjusted P < 0.01 and absolute log2(fold change) > 1. Principal component analysis and a heatmap of gene expression levels of each differentially expressed marker were analysed using the R prcomp function and hclust function (R version 3.6.1)The raw and processed RNA-seq data were deposited in the NCBI GEO database under the accession number GSE165620.
Chondrogenic induction
To induce chondrogenic differentiation under adherent culture conditions (2DCI), 2 × 104 cells were suspended in 100 µl ExpLBM medium containing 2 µl iMatrix511 and seeded into 96-well culture plates. After culturing for 3 d, the cells were washed with 1×PBS(−) and treated with chondrogenic culture step 1 medium (CDM2 medium + 3 µM CHIR99021 + 10 ng ml−1 FGF2 + 50 µg ml−1 ascorbic acid + 1×insulin-transferrin-selenium (ITS; Thermo Fisher) for 6 d. The step 1 medium was replaced with step 2 medium (CDM2 medium + 10 ng ml−1 FGF2 + 50 µg ml−1 ascorbic acid + 30 ng ml−1 BMP4 + 10 ng ml−1 TGF-β1 + 10 ng ml−1 GDF5 + 1×ITS) and the cells were cultured for 6 d. The culture media were replaced with fresh differentiation media every 3 d. To detect nodules on the culture dishes, the cells were fixed with 4% PFA for 20 min and then treated with 3% Alcian blue in 3% acetic acid for 30 min. After washing with 3% acetic acid, the Alcian blue-positive nodules were observed using a BZ-X710 microscope. To quantify the staining intensity, the cells were lysed with 6 M guanidinium chloride using a sonicator (Tomy) and the absorbance at 570 nm was measured using a Multiskan spectrophotometer. To induce chondrogenic induction using pellet culture conditions (3DCI), 1 × 105 ExpLBM cells were suspended in 200 µl chondrogenic culture step 1 medium and added to 96-well ultralow U-bottom plates (Corning). The cells were pelleted by centrifugation at 2,000 r.p.m. for 5 min. At the end of steps 1 and 2, the culture media were changed to chondrogenic culture step 2 and step 3 media (CDM2 medium + 50 µg ml−1 ascorbic acid + 30 ng ml−1 BMP4 + 10 ng ml−1 TGF-β1 + 10 ng ml−1 GDF5 + 1×ITS), respectively. The tissues were fixed with 4% PFA and the paraffin-embedded samples were cut into 4-µm-thick sections. The deparaffinized sections were stained with haematoxylin (20 min; Wako) and eosin (1 min; Wako). For the Alcian blue staining, the deparaffinized sections were stained with 3% Alcian blue or 3% acetic acid for 10 min and then washed with 3% acetic acid. For the Safranin O staining, the deparaffinized sections were stained with 0.05% fast green (Wako) solution for 5 min and then with 0.1% Safranin O (Wako) solution for 5 min. After staining, the samples were dehydrated and embedded with Entellan new reagent (Merck). Images were acquired using a BZ-X710 microscope.
Immunohistochemistry
Tissues were fixed with 4% PFA and the paraffin-embedded samples were cut into 4-µm-thick sections. The tissue samples were deparaffinized and the antigens were activated by heating the slides in 10 mM citrate buffer (pH 6.0). After treating the sections with 0.3% H2O2 in MeOH, the samples were incubated with blocking solution (3% normal goat serum and 0.1% Triton X-100 in 1×PBS(−)). The samples were treated with primary antibodies (diluted 1:200) for 1 h and then with secondary antibodies (diluted 1:400) for 1 h. The antibodies were diluted with blocking solution. After detecting signals with 3,3′-diaminobenzidine, the samples were dehydrated and embedded with Entellan new reagent (Merck). Images were acquired using a BZ-X710 microscope. The antibodies used are listed in Supplementary Table 8.
In vivo transplantation
The Okayama University Animal Care and Use Committee approved the experiments using mice and rats as well as the animal-care procedures. For subcutaneous transplantation, cartilaginous particles at step 3 (21 d) or ExpLBM cells with the βTCP scaffold (PENTAX) were transplanted into immunodeficient NOD-SCID mice (CLEA). After 4 or 8 weeks, the transplanted tissues were harvested and fixed with 4% PFA for 24 h. For transplantation of the cartilaginous particles, the skin and joint capsules of the knee joint of 10-week-old SCID rats (Tokyo University) were opened. A hole with a diameter of 1 mm and depth of 0.5 mm was drilled at the femoral groove. PRRX13′tdTomato-reporter-derived cartilaginous particles (step 3 treatment, 21 d) were cut in half and transplanted into the osteochondral defect of the articular cartilage via press fitting. The joint capsule and skin were closed and the rats were killed 4 weeks later. Tissue samples were paraffin-embedded at the Central Research Laboratory, Okayama University Medical School.
Transmission electron microscopy
Cartilaginous particles were fixed with 4% PFA and 2% glutaraldehyde and then fixed with 2% osmium tetroxide. After dehydration, embedding and polymerization, ultrathin sections were stained with 2% uranyl acetate and observed using a HITACHI H-7100S electron microscope at an acceleration voltage of 80 kV.
ExpLBM-based chemical screening
To induce chondrogenic differentiation under adherent culture conditions (2DCI), 2 × 104 ACGII-1-derived ExpLBM cells were suspended in 100 µl ExpLBM medium containing 2 µl iMatrix511 and seeded into 96-well culture plates. After culturing for 3 d, the cells were washed with 1×PBS(−) and treated with chondrogenic culture step 1 medium for 6 d. Chemical libraries (BML-2865 (Stock; 10 mM) and BML-2843J (Stock; 10 mM)) were provided by A. Hirao (Cancer Research Institute of Kanazawa University), and each chemical was diluted 2,000-fold in step 2 medium using multiple dispensers (SDR384SR, BioTec). The step 1 medium was then replaced with step 2 medium containing the appropriate chemical and the cells were cultured for 6 d. The culture media were replaced with fresh differentiation media every 3 d. To measure the nodule area or calculate the chondrogenic indexes of each well, phase-contrast images were taken using a BZ-X710 microscope and analysed using the BZ-X analyser software (Keyence). We defined the chondrogenic index as the nodule area of the chemical well or the control (empty) well. Chemicals with a chondrogenic index value >2 were selected for further analyses.
Zwitterionic copolymer-coated dish for mass production of cartilaginous particles
Iwai et al.34,35,36 reported cell self-aggregating, inducible specific charged culture surfaces, which are prepared by coating surfaces with zwitterionic copolymers comprising N,N-dimethylaminoethyl methacrylate (DMAEMA) and methacrylic acid (MA; PDMAEMA-co-PMA; Mn, 9.7 × 104 g mol−1; PDMAEMA/PMA ratio, 10). The cytotoxicity of this polymer was confirmed to be negative using the Ames test or dissolution test. Ultralow-attachment dishes (3.5 cm; PrimeSurface, Sumitomo Bakelite) were coated with droplets of zwitterionic copolymers at intervals of 0.5 mm using a droplet spotter (Biospot BT6000, MicroJet; 40 nl per spot, 300 spots per dish). Cells (1 × 106) were suspended in 1 ml ExpLBM medium and added to the polymer droplet-coated dishes. After incubation for 3 h, the culture media were replaced with fresh ExpLBM media and the cells were cultured for 2 d. The aggregates were then stripped using chondrogenic culture step 1 medium with 0.04% SphereMAX (Nisan Chemical) and transferred to six-well ultralow-attachment culture plates (Corning). When steps 1 and 2 were complete, the culture media were changed to chondrogenic culture step 2 media with 0.04% SphereMAX and step 3 media with 0.04% SphereMAX, respectively.
Statistical analysis
Data were analysed using Prism 9. All data were acquired by performing biological replicates of two or three independent experiments and are presented as the mean ± s.e.m. Statistical significance was determined using a two-tailed Student’s t-test and unpaired one-way or two-way analysis of variance (ANOVA) with correction for multiple testing using the Bonferroni method.
Study approval
The Ethics Committee of Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences approved the experimental protocols for studies of human participants. Written informed consent was provided by each donor.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and processed RNA-seq data were deposited in the NCBI GEO database under the accession number GSE165620. All data generated in this study, including source data and the data used to make the figures, are available from figshare with the identifier https://doi.org/10.6084/m9.figshare.14888937. Source data are provided with this paper.
Change history
05 December 2025
This article was originally published under standard Springer Nature license (© The Author(s), under exclusive licence to Springer Nature Limited). It is now available as an open-access paper under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International license, © The Author(s). The copyright has been amended in the HTML and PDF versions of the article.
References
Tam, W. L., Luyten, F. P. & Roberts, S. J. From skeletal development to the creation of pluripotent stem cell-derived bone-forming progenitors. Phil. Trans. R. Soc. B https://doi.org/10.1098/rstb.2017.0218 (2018).
Berendsen, A. D. & Olsen, B. R. Bone development. Bone 80, 14–18 (2015).
Kawata, M. et al. Simple and robust differentiation of human pluripotent stem cells toward chondrocytes by two small-molecule compounds. Stem Cell Rep. 13, 530–544 (2019).
Yamashita, A. et al. Generation of scaffoldless hyaline cartilaginous tissue from human iPSCs. Stem Cell Rep. 4, 404–418 (2015).
Craft, A. M. et al. Generation of articular chondrocytes from human pluripotent stem cells. Nat. Biotechnol. 33, 638–645 (2015).
Umeda, K. et al. Long-term expandable SOX9+ chondrogenic ectomesenchymal cells from human pluripotent stem cells. Stem. Cell Rep. 4, 712–726 (2015).
Nakajima, T. et al. Modeling human somite development and fibrodysplasia ossificans progressiva with induced pluripotent stem cells. Development https://doi.org/10.1242/dev.165431 (2018).
Tanaka, M. Molecular and evolutionary basis of limb field specification and limb initiation. Dev. Growth Differ. 55, 149–163 (2013).
Pignatti, E., Zeller, R. & Zuniga, A. To BMP or not to BMP during vertebrate limb bud development. Semin. Cell Dev. Biol. 32, 119–127 (2014).
Yu, K. & Ornitz, D. M. FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development 135, 483–491 (2008).
Lewandowski, J. P. et al. Spatiotemporal regulation of GLI target genes in the mammalian limb bud. Dev. Biol. 406, 92–103 (2015).
Taher, L. et al. Global gene expression analysis of murine limb development. PLoS ONE https://doi.org/10.1371/journal.pone.0028358 (2011).
Martin, J. F., Bradley, A. & Olson, E. N. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 9, 1237–1249 (1995).
ten Berge, D. et al. Prx1 and Prx2 are upstream regulators of sonic hedgehog and control cell proliferation during mandibular arch morphogenesis. Development 128, 2929–2938 (2001).
Dasouki, M., Andrews, B., Parimi, P. & Kamnasaran, D. Recurrent agnathia-otocephaly caused by DNA replication slippage in PRRX1. Am. J. Med. Genet. A 161A, 803–808 (2013).
Conway, S. J., Henderson, D. J. & Copp, A. J. Pax3 is required for cardiac neural crest migration in the mouse: evidence from the splotch (Sp2H) mutant. Development 124, 505–514 (1997).
Akiyama, H. et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl Acad. Sci. USA 102, 14665–14670 (2005).
Takarada, T. et al. Genetic analysis of Runx2 function during intramembranous ossification. Development 143, 211–218 (2016).
Durland, J. L., Sferlazzo, M., Logan, M. & Burke, A. C. Visualizing the lateral somitic frontier in the Prx1Cre transgenic mouse. J. Anat. 212, 590–602 (2008).
Karamboulas, K., Dranse, H. J. & Underhill, T. M. Regulation of BMP-dependent chondrogenesis in early limb mesenchyme by TGFβ signals. J. Cell Sci. 123, 2068–2076 (2010).
Tam, P. P. & Beddington, R. S. The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis. Development 99, 109–126 (1987).
Lawson, K. A., Meneses, J. J. & Pedersen, R. A. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113, 891–911 (1991).
Loh, K. M. et al. Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell 166, 451–467 (2016).
ten Berge, D., Brugmann, S. A., Helms, J. A. & Nusse, R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development 135, 3247–3257 (2008).
Hill, T. P., Taketo, M. M., Birchmeier, W. & Hartmann, C. Multiple roles of mesenchymal β-catenin during murine limb patterning. Development 133, 1219–1229 (2006).
Michos, O. et al. Gremlin-mediated BMP antagonism induces the epithelial–mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development 131, 3401–3410 (2004).
Stott, N. S. & Chuong, C. M. Dual action of sonic hedgehog on chondrocyte hypertrophy: retrovirus mediated ectopic sonic hedgehog expression in limb bud micromass culture induces cartilage nodules that are positive for alkaline phosphatase and type X collagen. J. Cell Sci. 110, 2691–2701 (1997).
Fernandez-Teran, M., Piedra, M. E., Rodriguez-Rey, J. C., Talamillo, A. & Ros, M. A. Expression and regulation of eHAND during limb development. Dev. Dyn. 226, 690–701 (2003).
Mahlapuu, M., Ormestad, M., Enerback, S. & Carlsson, P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development 128, 155–166 (2001).
Yang, L. et al. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development 133, 1575–1585 (2006).
Okada, M. et al. Modeling type II collagenopathy skeletal dysplasia by directed conversion and induced pluripotent stem cells. Hum. Mol. Genet. 24, 299–313 (2015).
Mortier, G. R. et al. Nosology and classification of genetic skeletal disorders: 2019 revision. Am. J. Med. Genet. A 179, 2393–2419 (2019).
Ala-Kokko, L., Baldwin, C. T., Moskowitz, R. W. & Prockop, D. J. Single base mutation in the type II procollagen gene (COL2A1) as a cause of primary osteoarthritis associated with a mild chondrodysplasia. Proc. Natl Acad. Sci. USA 87, 6565–6568 (1990).
Iwai, R., Nemoto, Y. & Nakayama, Y. The effect of electrically charged polyion complex nanoparticle-coated surfaces on adipose-derived stromal progenitor cell behaviour. Biomaterials 34, 9096–9102 (2013).
Iwai, R., Nemoto, Y. & Nakayama, Y. Preparation and characterization of directed, one-day-self-assembled millimeter-size spheroids of adipose-derived mesenchymal stem cells. J. Biomed. Mater. Res. A 104, 305–312 (2016).
Iwai, R., Haruki, R., Nemoto, Y. & Nakayama, Y. Induction of cell self-organization on weakly positively charged surfaces prepared by the deposition of polyion complex nanoparticles of thermoresponsive, zwitterionic copolymers. J. Biomed. Mater. Res. B 105, 1009–1015 (2017).
Schneider, V. A. & Mercola, M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 15, 304–315 (2001).
Mori, S. et al. Self-organized formation of developing appendages from murine pluripotent stem cells. Nat. Commun. 10, 3802 (2019).
Chen, Y., Xu, H. & Lin, G. Generation of iPSC-derived limb progenitor-like cells for stimulating phalange regeneration in the adult mouse. Cell Discov. 3, 17046 (2017).
Skinner, M. K. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res. C 93, 51–55 (2011).
Mohammad, H. P., Barbash, O. & Creasy, C. L. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat. Med. 25, 403–418 (2019).
Mazzone, R. et al. The emerging role of epigenetics in human autoimmune disorders. Clin. Epigenetics 11, 34 (2019).
Feinberg, A. P. The key role of epigenetics in human disease prevention and mitigation. N. Engl. J. Med. 378, 1323–1334 (2018).
Jarrell, D. K., Lennon, M. L. & Jacot, J. G. Epigenetics and mechanobiology in heart development and congenital heart disease. Diseases https://doi.org/10.3390/diseases7030052 (2019).
Lin, P. P. et al. Targeted mutation of p53 and Rb in mesenchymal cells of the limb bud produces sarcomas in mice. Carcinogenesis 30, 1789–1795 (2009).
Quist, T. et al. The impact of osteoblastic differentiation on osteosarcomagenesis in the mouse. Oncogene 34, 4278–4284 (2015).
Lee, J. Y. et al. Pre-transplantational control of the post-transplantational fate of human pluripotent stem cell-derived cartilage. Stem Cell Rep. 11, 440–453 (2018).
Reinisch, A. et al. Epigenetic and in vivo comparison of diverse MSC sources reveals an endochondral signature for human hematopoietic niche formation. Blood 125, 249–260 (2015).
Chamberlain, G., Fox, J., Ashton, B. & Middleton, J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells 25, 2739–2749 (2007).
Kim, M. et al. Donor variation and optimization of human mesenchymal stem cell chondrogenesis in hyaluronic acid. Tissue Eng. A 24, 1693–1703 (2018).
Garcia, J. et al. Chondrogenic potency analyses of donor-Matched chondrocytes and mesenchymal stem cells derived from bone marrow, infrapatellar fat pad, and subcutaneous fat. Stem Cells Int. 2016, 6969726 (2016).
Payne, K. A., Didiano, D. M. & Chu, C. R. Donor sex and age influence the chondrogenic potential of human femoral bone marrow stem cells. Osteoarthr. Cartil. 18, 705–713 (2010).
Acknowledgements
We thank N. Tsumaki for providing iPS cell lines derived from patients with COL2pathy (ACGII-1 and HCG-1); M. Hada and M. Nishio for technical assistance with feeder-free cultures; A. Hirao for providing chemical libraries; and K. Sekiguchi, S. Nagata, S. Tamaki, Y. Jin and C. Alev for their invaluable comments and advice. Preparation of slides and staining was supported by the Central Research Laboratory, Okayama University Medical School. We thank the members of the Department of Animal Resources, Advanced Science Research Center and Okayama University for maintaining the mice. This research was supported by Grants-in-aid for Scientific Research from the Japan Society for the Promotion of Science (grant no. 17H04399 to T. Takarada), AMED (grant nos 18bm0704024h0001 and 20bm0404064h0001 to T. Takarada), the Acceleration Program for Intractable Disease Research Utilizing Disease Specific iPS Cells (AMED) to J.T. and the Cooperative Research Program (Joint Usage/Research Center program) of the Institute for Frontier Life and Medical Sciences, Kyoto University to T. Takarada and J.T. These funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
D.Y. performed the experiments, analysed the data and wrote the manuscript. M.N., T. Takao, S.T., A.Y., S.K., A.M. and L.M. performed the experiments. H.Y., M.G., K.O., N.K. and T. Takarada provided critical materials. H.H., R.I., E.N., T.O. and J.T. discussed the data and provided critical advice. T. Takarada supervised the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biomedical Engineering thanks Shuibing Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Immunostaining of PRRX1 in human pluripotent stem cells on pluripotency (day 0) or in the PRRX1+ state.
Immunostaining of PRRX1 in hiPSC lines (414C2 and 1383D2) or hESC lines SEES4 and SEES5 at days 0 and 4.
Extended Data Fig. 2 Engraftment of cartilaginous particles in damaged articular cartilage of SCID rats.
A 1-mm drill hole defect was formed in the articular cartilage of SCID rats and then PRRX13´tdTomato-reporter-derived cartilaginous particles were embedded. The knee joints were fixed 4 weeks after transplantation and tissue sections were stained with HE, Toluidine blue or human VIMENTIN. Representative images were shown.
Extended Data Fig. 3 Bone-like-tissue forming capacity of ExpLBM cells.
(a-c) Human bone marrow-derived mesenchymal stem cell (BM-hMSC) or ExpLBM cells were suspended with βTCP and subcutaneously transplanted into NOD-SCID mice. After 8 weeks, mice were sacrificed and tissues were isolated to identify histological differences using HE staining (a), trichrome staining (b) and immunostaining with human VIMENTIN/COLX (c).
Extended Data Fig. 4 Abnormal chondrogenic capacity of ACGII-1- and HCG-1-derived ExpLBM cells.
(a) Alcian blue staining of other ACGII-1 or HCG-1 clone-derived ExpLBM cells after 2DCI. Other clones established from the same patient were induced to ExpLBM cells and their chondrogenic potentials were assessed by Alcian blue staining after 2DCI (n = 3, three biologically independent experiments). (b) Histological analysis of the cartilaginous particles generated using 3DCI. The cartilaginous particles were generated from each ExpLBM cell. Particle sections were stained with HE, Alcian blue or Safranin O or with antibodies against SOX9, COL2, COL1 or COLX. (c) Measurements and comparison of Safranin O-, SOX9-, or COL2-stained areas in Extended Data Fig. 5b (n = 3, three biologically independent experiments). (d) RT–qPCR analysis of the expression of chondrocyte marker genes after 3DCI using cartilaginous particles derived from each ExpLBM cell. All the values were normalized to the ACTB mRNA levels (n = 3, three biologically independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001. Statistical significance was determined by one-way ANOVA with correction for multiple testing using the Bonferroni method (a, c, d).
Extended Data Fig. 5 Transmission electron microscopy of cartilaginous particles of each ExpLBM cell on day 42 of step 3.
Cartilaginous particles of COL2pathy-ExpLBM cells comprise chondrocytes with a distended endoplasmic reticulum and low abundance of glycogen. Arrows and arrowheads indicate the endoplasmic reticulum (ER) and glycogen, respectively.
Supplementary information
Source data
Source data for Fig. 1
Unprocessed gels.
Source data for Fig. 1
Source data.
Source data for Fig. 3
Source data.
Source data for Fig. 4
Source data.
Source data for Fig. 5
Source data.
Source data for Extended Data Fig. 4
Source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yamada, D., Nakamura, M., Takao, T. et al. Induction and expansion of human PRRX1+ limb-bud-like mesenchymal cells from pluripotent stem cells. Nat Biomed Eng 5, 926–940 (2021). https://doi.org/10.1038/s41551-021-00778-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41551-021-00778-x
This article is cited by
-
Generation and transcriptome profiling of bonobo induced pluripotent stem cells using stealth RNA vectors: a tripartite comparative study with humans and chimpanzees
BMC Genomics (2025)
-
Recapitulation of endochondral ossification by hPSC-derived SOX9+ sclerotomal progenitors
Nature Communications (2025)
-
Bioengineered chondrocyte-products from human induced pluripotent stem cells are useful for repairing articular cartilage injury in minipig model
npj Regenerative Medicine (2025)
-
In situ cell condensation-based cartilage tissue engineering via immediately implantable high-density stem cell core and rapidly degradable shell microgels
Advanced Composites and Hybrid Materials (2025)
-
Effect of a retinoic acid analogue on BMP-driven pluripotent stem cell chondrogenesis
Scientific Reports (2024)