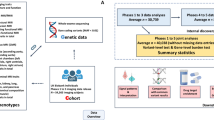
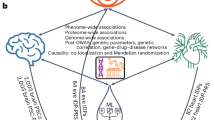
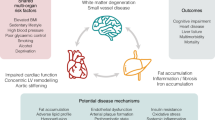
Abstract
Understanding the complex relationships among major clinical outcomes and the interplay among multiple organs remains a considerable challenge. By using imaging phenotypes, we can characterize the functional and structural architecture of major human organs. Mendelian randomization (MR) provides a valuable framework for uncovering robust relationships between phenotypes by leveraging genetic variants as instrumental variables. Here we conduct a systematic multi-organ MR analysis involving 402 imaging traits and 372 clinical outcomes. Our analysis reveals 184 MR associations for 58 diseases and 56 imaging traits across various organs, tissues and systems, including the brain, heart, liver, kidney, lung, pancreas, spleen, adipose tissue and skeletal system. We identify intra-organ MR connections, such as the putative bidirectional genetic links between Alzheimer’s disease and brain function, and interorgan associations, such as heart diseases and brain health. Metabolic disorders, such as diabetes, show genetically rooted putative MR effects across multiple organs. These findings shed light on the genetic links spanning multiple organs, providing targets for future mechanistic follow-up for clinical disease research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
We used summary-level GWAS data in this study, which can be obtained from the FinnGen project (https://www.finngen.fi/en/access_results), BIG-KP (https://bigkp.org/) and Heart-KP (https://heartkp.org/), and project-specific resources are detailed in refs. 3 and 8.
Code availability
We used publicly available software and tools. Our analysis code is available on Zenodo at https://doi.org/10.5281/zenodo.16518650 (ref. 149).
References
Buckner, R. L. et al. Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory. J. Neurosci. 25, 7709–7717 (2005).
Pennell, D. J. et al. Clinical indications for cardiovascular magnetic resonance (CMR): consensus panel report. Eur. Heart J. 25, 1940–1965 (2004).
Kun, E. et al. The genetic architecture and evolution of the human skeletal form. Science 381, eadf8009 (2023).
Petersen, S. E. et al. UK Biobank’s cardiovascular magnetic resonance protocol. J. Cardiovasc. Magn. Reson. 18, 8 (2015).
Littlejohns, T. J., Sudlow, C., Allen, N. E. & Collins, R. UK Biobank: opportunities for cardiovascular research. Eur. Heart J. 40, 1158–1166 (2019).
Miller, K. L. et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 19, 1523–1536 (2016).
Thompson, P.M. et al. ENIGMA and global neuroscience: a decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl. Psychiatry 10, 100 (2020).
Liu, Y. et al. Genetic architecture of 11 organ traits derived from abdominal MRI using deep learning. eLife 10, e65554 (2021).
Smith, S. M. & Nichols, T. E. Statistical challenges in ‘big data’ human neuroimaging. Neuron 97, 263–268 (2018).
Tian, Y. E. et al. Heterogeneous aging across multiple organ systems and prediction of chronic disease and mortality. Nat. Med. 29, 1221–1231 (2023).
Taschler, B., Smith, S.M. & Nichols, T.E. Causal inference on neuroimaging data with Mendelian randomisation. NeuroImage 258, 119385 (2022).
Sanderson, E. et al. Mendelian randomization. Nat. Rev. Methods Prim. 2, 6 (2022).
Pingault, J.-B. et al. Using genetic data to strengthen causal inference in observational research. Nat. Rev. Genet. 19, 566–580 (2018).
Aung, N. et al. Genome-wide analysis of left ventricular image-derived phenotypes identifies fourteen loci associated with cardiac morphogenesis and heart failure development. Circulation 140, 1318–1330 (2019).
Córdova-Palomera, A. et al. Cardiac imaging of aortic valve area from 34,287 UK Biobank participants reveals novel genetic associations and shared genetic comorbidity with multiple disease phenotypes. Circ. Genom. Precis. Med. 13, e003014 (2020).
Meyer, H. V. et al. Genetic and functional insights into the fractal structure of the heart. Nature 584, 589–594 (2020).
Pirruccello, J. P. et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nat. Commun. 11, 2254 (2020).
Pirruccello, J. P. et al. Genetic analysis of right heart structure and function in 40,000 people. Nat. Genet. 54, 792–803 (2022).
Thanaj, M. et al. Genetic and environmental determinants of diastolic heart function. Nat. Cardiovasc. Res. 1, 361–371 (2022).
Elliott, L. T. et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 210–216 (2018).
Zhao, B. et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat. Genet. 51, 1637–1644 (2019).
Smith, S. M. et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat. Neurosci. 24, 737–745 (2021).
Zhao, B. et al. Common genetic variation influencing human white matter microstructure. Science 372, eabf3736 (2021).
Grasby, K. L. et al. The genetic architecture of the human cerebral cortex. Science 367, eaay6690 (2020).
Zhao, B. et al. Genetic influences on the intrinsic and extrinsic functional organizations of the cerebral cortex. Preprint at medRxiv https://doi.org/10.1101/2021.07.27.21261187 (2021).
Watanabe, K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51, 1339–1348 (2019).
Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023).
Flynn, B. I. et al. Deep learning based phenotyping of medical images improves power for gene discovery of complex disease. npj Digit. Med. 6, 155 (2023).
Guo, J. et al. Mendelian randomization analyses support causal relationships between brain imaging-derived phenotypes and risk of psychiatric disorders. Nat. Neurosci. 25, 1519–1527 (2022).
Chen, X. et al. Kidney damage causally affects the brain cortical structure: a Mendelian randomization study. eBioMedicine 72, 103592 (2021).
Williams, J. A. et al. Inflammation and brain structure in schizophrenia and other neuropsychiatric disorders: a Mendelian randomization study. JAMA Psychiatry 79, 498–507 (2022).
Topiwala, A. et al. Associations between moderate alcohol consumption, brain iron, and cognition in UK Biobank participants: observational and mendelian randomization analyses. PLoS Med. 19, e1004039 (2022).
Holmes, M. V. et al. Mendelian randomization of blood lipids for coronary heart disease. Eur. Heart J. 36, 539–550 (2015).
Lamina, C. & Kronenberg, F. Estimation of the required lipoprotein (a)-lowering therapeutic effect size for reduction in coronary heart disease outcomes: a Mendelian randomization analysis. JAMA Cardiol. 4, 575–579 (2019).
Skou, S. T. et al. Multimorbidity. Nat. Rev. Dis. Prim. 8, 48 (2022).
Langenberg, C., Hingorani, A. D. & Whitty, C. J. Biological and functional multimorbidity—from mechanisms to management. Nat. Med. 29, 1649–1657 (2023).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Bijsterbosch, J. et al. Investigations into within-and between-subject resting-state amplitude variations. NeuroImage 159, 57–69 (2017).
Bai, W. et al. A population-based phenome-wide association study of cardiac and aortic structure and function. Nat. Med. 26, 1654–1662 (2020).
Zhao, B. et al. Heart-brain connections: phenotypic and genetic insights from magnetic resonance images. Science 380, abn6598 (2023).
Bowden, J. et al. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 36, 1783–1802 (2017).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Bowden, J. et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int J. Epidemiol. 48, 728–742 (2019).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Hartwig, F. P., Davey Smith, G. & Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998 (2017).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Ye, T., Shao, J. & Kang, H. Debiased inverse-variance weighted estimator in two-sample summary-data Mendelian randomization. Ann. Stat. 49, 2079–2100 (2021).
Zhao, Q., Wang, J., Hemani, G., Bowden, J. & Small, D. S. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann. Stat. 48, 1742–1769 (2020).
Wang, J. et al. Causal inference for heritable phenotypic risk factors using heterogeneous genetic instruments. PLoS Genet. 17, e1009575 (2021).
Bennett, I. J., Madden, D. J., Vaidya, C. J., Howard, D. V. & Howard, J. H. Jr Age-related differences in multiple measures of white matter integrity: a diffusion tensor imaging study of healthy aging. Hum. Brain Mapp. 31, 378–390 (2010).
Cerqueira, M. D.et al.; American Heart Association Writing Group on Myocardial Segmentation and Registration for Cardiac Imaging Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart: a statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation 105, 539–542 (2002).
Stratos, C., Stefanadis, C., Kallikazaros, I., Boudoulas, H. & Toutouzas, P. Ascending aorta distensibility abnormalities in hypertensive patients and response to nifedipine administration. Am. J. Med. 93, 505–512 (1992).
Asmar, R. et al. Aortic distensibility in normotensive, untreated and treated hypertensive patients. Blood Press. 4, 48–54 (1995).
Nabati, M., Namazi, S. S., Yazdani, J. & Sharif Nia, H. Relation between aortic stiffness index and distensibility with age in hypertensive patients. Int. J. Gen. Med.13, 297–303 (2020).
Berman, M. N., Tupper, C. & Bhardwaj, A. in StatPearls (StatPearls Publishing, 2022).
Kim, D.-Y. & Camilleri, M. Serotonin: a mediator of the brain–gut connection. Am. J. Gastroenterol. 95, 2698–2709 (2000).
Jones, M., Dilley, J., Drossman, D. & Crowell, M. Brain–gut connections in functional GI disorders: anatomic and physiologic relationships. Neurogastroenterol. Motil. 18, 91–103 (2006).
Keefer, L. et al. A Rome working team report on brain–gut behavior therapies for disorders of gut–brain interaction. Gastroenterology 162, 300–315 (2022).
Xie, Z., Tong, S., Chu, X., Feng, T. & Geng, M. Chronic kidney disease and cognitive impairment: the kidney–brain axis. Kidney Dis. 8, 275–285 (2022).
de Donato, A., Buonincontri, V., Borriello, G., Martinelli, G. & Mone, P. The dopamine system: insights between kidney and brain. Kidney Blood Press. Res. 47, 493–505 (2022).
McCracken, C. et al. Multi-organ imaging demonstrates the heart–brain–liver axis in UK Biobank participants. Nat. Commun. 13, 7839 (2022).
Walker, V. M., Zheng, J., Gaunt, T. R. & Smith, G. D. Phenotypic causal inference using genome-wide association study data: Mendelian randomization and beyond. Annu. Rev. Biomed. Data Sci. 5, 1–17 (2022).
Burgess, S., Davies, N. M. & Thompson, S. G. Bias due to participant overlap in two-sample Mendelian randomization. Genet. Epidemiol. 40, 597–608 (2016).
Jaggi, A. et al. A structural heart–brain axis mediates the association between cardiovascular risk and cognitive function. Imaging Neurosci. 2, imag-2-00063 (2024).
Berridge, K. C. & Kringelbach, M. L. Pleasure systems in the brain. Neuron 86, 646–664 (2015).
Bressler, S. L. & Menon, V. Large-scale brain networks in cognition: emerging methods and principles. Trends Cogn. Sci. 14, 277–290 (2010).
Wang, K. et al. Altered functional connectivity in early Alzheimer’s disease: a resting-state fMRI study. Hum. Brain Mapp. 28, 967–978 (2007).
Sorg, C. et al. Selective changes of resting-state networks in individuals at risk for Alzheimer’s disease. Proc. Natl Acad. Sci. USA 104, 18760–18765 (2007).
Ranasinghe, K. G. et al. Regional functional connectivity predicts distinct cognitive impairments in Alzheimer’s disease spectrum. NeuroImage Clin. 5, 385–395 (2014).
Pini, L. et al. A low-dimensional cognitive-network space in Alzheimer’s disease and frontotemporal dementia. Alzheimer’s Res. Ther. 14, 199 (2022).
Torso, M. et al. In vivo cortical diffusion imaging relates to Alzheimer’s disease neuropathology. Alzheimer’s Res. Ther. 15, 165 (2023).
Tu, M.-C. et al. Joint diffusional kurtosis magnetic resonance imaging analysis of white matter and the thalamus to identify subcortical ischemic vascular disease. Sci. Rep. 14, 2570 (2024).
Liu, W. et al. Brain–heart communication in health and diseases. Brain Res. Bull. 183, 27–37 (2022).
Walker, K. A., Power, M. C. & Gottesman, R. F. Defining the relationship between hypertension, cognitive decline, and dementia: a review. Curr. Hypertens. Rep. 19, 24 (2017).
Zhang, H. et al. Reduced regional gray matter volume in patients with chronic obstructive pulmonary disease: a voxel-based morphometry study. Am. J. Neuroradiol. 34, 334–339 (2013).
Yang, C., Hawkins, K. E., Doré, S. & Candelario-Jalil, E. Neuroinflammatory mechanisms of blood–brain barrier damage in ischemic stroke. Am. J. Physiol. Cell Physiol. 316, C135–C153 (2019).
Carnevale, D. et al. Role of neuroinflammation in hypertension-induced brain amyloid pathology. Neurobiol. Aging 33, 205.e219–205.e229 (2012).
Haspula, D. & Clark, M. A. Neuroinflammation and sympathetic overactivity: mechanisms and implications in hypertension. Auton. Neurosci. 210, 10–17 (2018).
Sweeney, M. D., Sagare, A. P. & Zlokovic, B. V. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 14, 133–150 (2018).
Niedermeyer, E. Alzheimer disease: caused by primary deficiency of the cerebral blood flow. Clin. EEG Neurosci. 37, 175–177 (2006).
Kisler, K., Nelson, A. R., Montagne, A. & Zlokovic, B. V. Cerebral blood flow regulation and neurovascular dysfunction in Alzheimer disease. Nat. Rev. Neurosci. 18, 419–434 (2017).
Chu, B., Marwaha, K., Sanvictores, T. & Ayers, D. in StatPearls (StatPearls Publishing, 2021).
Charmandari, E., Tsigos, C. & Chrousos, G. Endocrinology of the stress response. Annu. Rev. Physiol. 67, 259–284 (2005).
Colao, A., Marzullo, P., Di Somma, C. & Lombardi, G. Growth hormone and the heart. Clin. Endocrinol. 54, 137–154 (2001).
Fazio, S. et al. Growth hormone and heart performance: a novel mechanism of cardiac wall stress regulation in humans. Eur. Heart J. 18, 340–347 (1997).
Black, P. H. & Garbutt, L. D. Stress, inflammation and cardiovascular disease. J. Psychosom. Res. 52, 1–23 (2002).
Libby, P. Inflammation and cardiovascular disease mechanisms. Am. J. Clin. Nutr. 83, 456S–460S (2006).
Holmes, C. Systemic inflammation and A lzheimer’s disease. Neuropathol. Appl. Neurobiol. 39, 51–68 (2013).
Laleman, W., Claria, J., Van der Merwe, S., Moreau, R. & Trebicka, J. Systemic inflammation and acute-on-chronic liver failure: too much, not enough. Can. J. Gastroenterol. Hepatol. 2018, 1027152 (2018).
Scherder, E. J., Bogen, T., Eggermont, L. H., Hamers, J. P. & Swaab, D. F. The more physical inactivity, the more agitation in dementia. Int. Psychogeriatr. 22, 1203–1208 (2010).
Peckett, A. J., Wright, D. C. & Riddell, M. C. The effects of glucocorticoids on adipose tissue lipid metabolism. Metabolism 60, 1500–1510 (2011).
Polkey, M. I., Lyall, R. A., Moxham, J. & Leigh, P. N. Respiratory aspects of neurological disease. J. Neurol. Neurosurg. Psychiatry 66, 5–15 (1999).
Pollock, R. D., Rafferty, G. F., Moxham, J. & Kalra, L. Respiratory muscle strength and training in stroke and neurology: a systematic review. Int. J. Stroke 8, 124–130 (2013).
Kushner, T. & Cafardi, J. Chronic liver disease and COVID-19: alcohol use disorder/alcohol-associated liver disease, nonalcoholic fatty liver disease/nonalcoholic steatohepatitis, autoimmune liver disease, and compensated cirrhosis. Clin. Liver Dis. 15, 195 (2020).
Rhyou, H.-I. & Nam, Y.-H. Association between cognitive function and asthma in adults. Ann. Allergy Asthma Immunol. 126, 69–74 (2021).
Ray, M., Sano, M., Wisnivesky, J. P., Wolf, M. S. & Federman, A. D. Asthma control and cognitive function in a cohort of elderly adults. J. Am. Geriatrics Soc. 63, 684–691 (2015).
Alvarez, J. I., Cayrol, R. & Prat, A. Disruption of central nervous system barriers in multiple sclerosis. Biochimic. Biophys. Acta 1812, 252–264 (2011).
Krupp, L. B. et al. International Pediatric Multiple Sclerosis Study Group criteria for pediatric multiple sclerosis and immune-mediated central nervous system demyelinating disorders: revisions to the 2007 definitions. Mult. Scler. J. 19, 1261–1267 (2013).
Huda, S. et al. Neuromyelitis optica spectrum disorders. Clin. Med. 19, 169 (2019).
Kim, W., Kim, S.-H., Huh, S.-Y. & Kim, H. J. Brain abnormalities in neuromyelitis optica spectrum disorder. Mult. Scler. Int. 2012, 735486 (2012).
Lancaster, E. The diagnosis and treatment of autoimmune encephalitis. J. Clin. Neurol. 12, 1–13 (2016).
Wartolowska, K. et al. Structural changes of the brain in rheumatoid arthritis. Arthritis Rheum. 64, 371–379 (2012).
Kozora, E. & Filley, C. M. Cognitive dysfunction and white matter abnormalities in systemic lupus erythematosus. J. Int. Neuropsychol. Soc. 17, 385–392 (2011).
Appenzeller, S. et al. Longitudinal analysis of gray and white matter loss in patients with systemic lupus erythematosus. NeuroImage 34, 694–701 (2007).
Rosenberg, G. A. Inflammation and white matter damage in vascular cognitive impairment. Stroke 40, S20–S23 (2009).
Raj, D. et al. Increased white matter inflammation in aging-and Alzheimer’s disease brain. Front. Mol. Neurosci. 10, 206 (2017).
Gerdts, E. et al. Correlates of left atrial size in hypertensive patients with left ventricular hypertrophy: the Losartan Intervention For Endpoint Reduction in Hypertension (LIFE) Study. Hypertension 39, 739–743 (2002).
Eshoo, S., Ross, D. L. & Thomas, L. Impact of mild hypertension on left atrial size and function. Circ. Cardiovasc. Imaging 2, 93–99 (2009).
Sanfilippo, A. J. et al. Atrial enlargement as a consequence of atrial fibrillation. A prospective echocardiographic study. Circulation 82, 792–797 (1990).
Saheera, S. & Krishnamurthy, P. Cardiovascular changes associated with hypertensive heart disease and aging. Cell Transplant. 29, 963689720920830 (2020).
Hiraiwa, H. et al. Clinical significance of spleen size in patients with heart failure. Eur. Heart J. 42, ehab724.0756 (2021).
Ormazabal, V. et al. Association between insulin resistance and the development of cardiovascular disease. Cardiovasc. Diabetol. 17, 122 (2018).
Shah, A., Mehta, N. & Reilly, M. P. Adipose inflammation, insulin resistance, and cardiovascular disease. J. Parenter. Enter. Nutr. 32, 638–644 (2008).
Boudina, S. & Abel, E. D. Diabetic cardiomyopathy, causes and effects. Rev. Endocr. Metab. Disord. 11, 31–39 (2010).
Horton, W. B. & Barrett, E. J. Microvascular dysfunction in diabetes mellitus and cardiometabolic disease. Endocr. Rev. 42, 29–55 (2021).
Kibel, A. et al. Coronary microvascular dysfunction in diabetes mellitus. J. Int. Med. Res. 45, 1901–1929 (2017).
Fuentes-Antrás, J. et al. Targeting metabolic disturbance in the diabetic heart. Cardiovasc. Diabetol. 14, 17 (2015).
Wagner, R. et al. Metabolic implications of pancreatic fat accumulation. Nat. Rev. Endocrinol. 18, 43–54 (2022).
Yaney, G. C. & Corkey, B. E. Fatty acid metabolism and insulin secretion in pancreatic beta cells. Diabetologia 46, 1297–1312 (2003).
Dludla, P. V. et al. Pancreatic beta-cell dysfunction in type 2 diabetes: implications of inflammation and oxidative stress. World J. Diabetes 14, 130–146 (2023).
Kocaturk, E., Kar, E., Kusku Kiraz, Z. & Alatas, O. Insulin resistance and pancreatic beta cell dysfunction are associated with thyroid hormone functions: a cross-sectional hospital-based study in Turkey. Diabetes Metab. Syndr. 14, 2147–2151 (2020).
Meeks, K. A. C., Adeyemo, A. & Agyemang, C. Beta-cell dysfunction and insulin resistance in relation to abnormal glucose tolerance in African populations: can we afford to ignore the diversity within African populations? BMJ Open Diabetes Res. Care 10, e002685 (2022).
Bonora, E. et al. Insulin resistance and beta-cell dysfunction in newly diagnosed type 2 diabetes: expression, aggregation and predominance. Verona Newly Diagnosed Type 2 Diabetes Study 10. Diabetes Metab. Res Rev. 38, e3558 (2022).
Whalen, R., Carter, D. & Steele, C. Influence of physical activity on the regulation of bone density. J. Biomech. 21, 825–837 (1988).
Sanderson, E., Spiller, W. & Bowden, J. Testing and correcting for weak and pleiotropic instruments in two-sample multivariable Mendelian randomization. Stat. Med. 40, 5434–5452 (2021).
Zhao, Q., Wang, J., Spiller, W., Bowden, J. & Small, D. S. Two-sample instrumental variable analyses using heterogeneous samples. Stat. Sci. 34, 317–333 (2019).
Cui, R. et al. Improving fine-mapping by modeling infinitesimal effects. Nat. Genet. 56, 162–169 (2024).
Xue, H., Shen, X. & Pan, W. Causal inference in transcriptome-wide association studies with invalid instruments and GWAS summary data. J. Am. Stat. Assoc.118, 1525–1537 (2023).
Hu, X. et al. Benchmarking Mendelian randomization methods for causal inference using genome-wide association study summary statistics. Am. J. Hum. Genet 111, 1717–1735 (2024).
Richmond, R. C. & Smith, G. D. Mendelian randomization: concepts and scope. Cold Spring Harb. Perspect. Med. 12, a040501 (2022).
Tseng, W. Y., Su, M. Y. & Tseng, Y. H. Introduction to cardiovascular magnetic resonance: technical principles and clinical applications. Acta Cardiol. Sin. 32, 129–144 (2016).
Pennell, D. J. Cardiovascular magnetic resonance. Circulation 121, 692–705 (2010).
Bai, W. et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 20, 65 (2018).
Bai, W. et al. Recurrent neural networks for aortic image sequence segmentation with sparse annotations. In Proc. Medical Image Computing and Computer Assisted Intervention—MICCAI 2018 (eds Frangi, A. et al.) 586–594 (2018).
Zhao, B. et al. Heritability of regional brain volumes in large-scale neuroimaging and genetic studies. Cereb. Cortex 29, 2904–2914 (2019).
Zhao, B. et al. Large-scale GWAS reveals genetic architecture of brain white matter microstructure and genetic overlap with cognitive and mental health traits (n = 17,706). Mol. Psychiatry 26, 3943–3955 (2021).
Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54, 2033–2044 (2011).
Jahanshad, N. et al. Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: a pilot project of the ENIGMA-DTI working group. NeuroImage 81, 455–469 (2013).
Kochunov, P. et al. Multi-site study of additive genetic effects on fractional anisotropy of cerebral white matter: comparing meta and megaanalytical approaches for data pooling. NeuroImage 95, 136–150 (2014).
Glasser, M. F. et al. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016).
Ji, J. L. et al. Mapping the human brain’s cortical-subcortical functional network organization. NeuroImage 185, 35–57 (2019).
Deng, L., Zhang, H. & Yu, K. Power calculation for the general two-sample Mendelian randomization analysis. Genet Epidemiol. 44, 290–299 (2020).
Levy, D. et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 41, 677–687 (2009).
Burton, P. R.et al.; Wellcome Trust Case Control Consortium; Australo-Anglo-American Spondylitis Consortium (TASC) Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Schwartzentruber, J. et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat. Genet. 53, 392–402 (2021).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022).
Verma, A. et al. Diversity and scale: genetic architecture of 2,068 traits in the VA Million Veteran Program. Science 385, eadj1182 (2024).
Zhu, Z. et al. Genetic overlap of chronic obstructive pulmonary disease and cardiovascular disease-related traits: a large-scale genome-wide cross-trait analysis. Respir. Res 20, 64 (2019).
Shu, J. MOMR_code. Zenodo https://doi.org/10.5281/zenodo.16518650 (2025).
Acknowledgements
Research reported in this publication was supported by the National Institute of Mental Health under award number R01MH136055 (B.Z.) and National Institute on Aging under award numbers RF1AG082938 (B.Z. and H.Z.) and R01AG085581 (B.Z. and H.Z.). Assistance for this project was provided by the UNC Intellectual and Developmental Disabilities Research Center (NICHD; P50 HD103573; H.Z.), and by grants RF1AG098697 (H.Z.), R01AR082684 (H.Z.), OT2OD038045-01 (H.Z.) and K01AG095286 (T.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The study has also been partially supported by funding from the Purdue University Statistics Department, Department of Statistics and Data Science at the University of Pennsylvania, Wharton Dean’s Research Fund, Analytics at Wharton, Wharton AI and Analytics Initiative, Perelman School of Medicine CCEB Innovation Center Grant and the University Research Foundation Grant (B.Z.). This research has been conducted using summary-level data from the UKB study and the FinnGen research project. We thank the individuals who participated in the UKB and FinnGen studies for their contribution and the research teams for their efforts in collecting, processing and disseminating these datasets. We thank the research computing groups at the University of North Carolina at Chapel Hill, Purdue University and the Wharton School of the University of Pennsylvania for providing computational resources and support that have contributed to these research results.
Author information
Authors and Affiliations
Contributions
J.S. and B.Z. designed the study. J.S., C.C., B.L., Z.F., X.Y., Y.Y, X.W. and Y.L. analysed the data. R.Z. and J.C. helped interpret the findings. B.X., T.L. and H.Z. provided feedback on the results. J.S. and B.Z. wrote the paper with feedback from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Marios Georgakis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note, Figs. 1–14 and legends for Tables 1–17.
Supplementary Tables
Supplementary Tables 1–17 provide data descriptions and detailed MR results.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shu, J., Zheng, R., Chirinos, J. et al. Inferring multi-organ genetic connections using imaging and clinical data through Mendelian randomization. Nat. Biomed. Eng (2025). https://doi.org/10.1038/s41551-025-01554-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41551-025-01554-x