Abstract
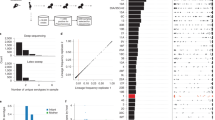
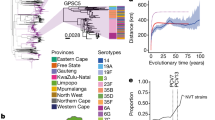
Many bacterial species are composed of multiple lineages distinguished by extensive variation in gene content. These often cocirculate in the same habitat, but the evolutionary and ecological processes that shape these complex populations are poorly understood. Addressing these questions is particularly important for Streptococcus pneumoniae, a nasopharyngeal commensal and respiratory pathogen, because the changes in population structure associated with the recent introduction of partial-coverage vaccines have substantially reduced pneumococcal disease. Here we show that pneumococcal lineages from multiple populations each have a distinct combination of intermediate-frequency genes. Functional analysis suggested that these loci may be subject to negative frequency-dependent selection (NFDS) through interactions with other bacteria, hosts or mobile elements. Correspondingly, these genes had similar frequencies in four populations with dissimilar lineage compositions. These frequencies were maintained following substantial alterations in lineage prevalences once vaccination programmes began. Fitting a multilocus NFDS model of post-vaccine population dynamics to three genomic datasets using Approximate Bayesian Computation generated reproducible estimates of the influence of NFDS on pneumococcal evolution, the strength of which varied between loci. Simulations replicated the stable frequency of lineages unperturbed by vaccination, patterns of serotype switching and clonal replacement. This framework highlights how bacterial ecology affects the impact of clinical interventions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Haegeman, B. & Weitz, J. S. A neutral theory of genome evolution and the frequency distribution of genes. BMC Genomics 13, 196 (2012).
Baumdicker, F., Hess, W. R. & Pfaffelhuber, P. The infinitely many genes model for the distributed genome of bacteria. Genome Biol. Evol. 4, 443–456 (2012).
Marttinen, P., Croucher, N. J., Gutmann, M. U., Corander, J. & Hanage W. P. Recombination produces coherent bacterial species clusters in both core and accessory genomes. Microb. Genom. 1, e000038 (2015).
Hogg, J. S. et al. Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 8, R103 (2007).
Collins, R. E. & Higgs, P. G. Testing the infinitely many genes model for the evolution of the bacterial core genome and pangenome. Mol. Biol. Evol. 29, 3413–3425 (2012).
Lobkovsky, A. E., Wolf, Y. I. & Koonin, E. V. Gene frequency distributions reject a neutral model of genome evolution. Genome Biol. Evol. 5, 233–242 (2013).
McInerney, J. O., McNally, A. & O’Connell, M. J. Why prokaryotes have pangenomes. Nat. Microbiol. 2, 17040 (2017).
Shapiro, B. J. et al. Population genomics of early events in the ecological differentiation of bacteria. Science 336, 48–51 (2012).
Cohan, F. M. Bacterial species and speciation. Syst. Biol. 50, 513–524 (2001).
Cohan, F. M. What are bacterial species? Annu. Rev. Microbiol. 56, 457–487 (2002).
Watkins, E. R. et al. Vaccination drives changes in metabolic and virulence profiles of Streptococcus pneumoniae. PLoS Pathog. 11, e1005034 (2015).
Regev-Yochay, G. et al. Re-emergence of the type 1 pilus among Streptococcus pneumoniae isolates in Massachusetts, USA. Vaccine 28, 4842–4846 (2010).
Cobey, S. & Lipsitch, M. Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science 335, 1376–1380 (2012).
Croucher, N. J. et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat. Genet. 45, 656–663 (2013).
Huang, S. S. et al. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics 124, e1–11 (2009).
Gladstone, R. A. et al. Five winters of pneumococcal serotype replacement in UK carriage following PCV introduction. Vaccine 33, 2015–2021 (2015).
Gladstone, R. A. et al. Pre-vaccine serotype composition within a lineage signposts its serotype replacement — a carriage study over 7 years following pneumococcal conjugate vaccine use in the UK. Microb. Genom. 3, 119 (2017).
Cremers, A. J. H. et al. The post-vaccine microevolution of invasive Streptococcus pneumoniae. Sci. Rep. 5, 14952 (2015).
Levin, B. R. Frequency-dependent selection in bacterial populations. Phil. Trans. R. Soc. Lond. B 319, 459–472 (1988).
Maynard Smith, J. Evolutionary Genetics 2nd edn (Oxford Univ. Press, Oxford, 1998).
Croucher, N. J. et al. Diversification of bacterial genome content through distinct mechanisms over different timescales. Nat. Commun. 5, 5471 (2014).
Croucher, N. J. et al. Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict. PLoS Biol. 14, e1002394 (2016).
Takeuchi, N., Cordero, O. X., Koonin, E. V. & Kaneko, K. Gene-specific selective sweeps in bacteria and archaea caused by negative frequency-dependent selection. BMC Biol. 13, 20 (2015).
Cordero, O. X. & Polz, M. F. Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 12, 263–273 (2014).
Dawid, S., Roche, A. M. & Weiser, J. N. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75, 443–451 (2007).
Miller, E. L., Abrudan, M. I., Roberts, I. S. & Rozen, D. E. Diverse ecological strategies are encoded by Streptococcus pneumoniae bacteriocin-like peptides. Genome Biol. Evol. 8, 1072–1090 (2016).
Bogaardt, C., van Tonder, A. J. & Brueggemann, A. B. Genomic analyses of pneumococci reveal a wide diversity of bacteriocins — including pneumocyclicin, a novel circular bacteriocin. BMC Genomics 16, 554 (2015).
Maricic, N., Anderson, E. S., Opipari, A. M. E., Yu, E. A. & Dawid, S. Characterization of a multipeptide lantibiotic locus in Streptococcus pneumoniae. mBio 7, e01656-15 (2016).
Hoover, S. E. et al. A new quorum-sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Mol. Microbiol. 97, 229–243 (2015).
Kerr, B., Riley, M. A., Feldman, M. W. & Bohannan, B. J. M. Local dispersal promotes biodiversity in a real-life game of rock–paper–scissors. Nature 418, 171–174 (2002).
Stewart, F. M. & Levin, B. R. Partitioning of resources and the outcome of interspecific competition: a model and some general considerations. Am. Nat. 107, 171–198 (1973).
Levin, B. R. Coexistence of two asexual strains on a single resource. Science 175, 1272–1274 (1972).
Colijn, C. & Cohen, T. How competition governs whether moderate or aggressive treatment minimizes antibiotic resistance. eLife 4, e10559 (2015).
Lehtinen, S. et al. Evolution of antibiotic resistance is linked to any genetic mechanism affecting bacterial duration of carriage. Proc. Natl Acad. Sci. USA 114, 1075–1080 (2017).
Croucher, N. J. et al. Diverse evolutionary patterns of pneumococcal antigens identified by pangenome-wide immunological screening. Proc. Natl Acad. Sci. USA 114, E357–E366 (2017).
Croucher, N. J. et al. Selective and genetic constraints on pneumococcal serotype switching. PLoS Genet. 11, e1005095 (2015).
Bagnoli, F. et al. A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J. Bacteriol. 190, 5480–5492 (2008).
Chewapreecha, C. et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 46, 305–309 (2014).
Goossens, H. et al. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365, 579–587 (2005).
Gutmann, M. U. & Corander, J. Bayesian optimization for likelihood-free inference of simulator-based statistical models. J. Mach. Learn. Res. 17, 1–47 (2016).
Lintusaari, J., Gutmann, M. U., Dutta, R., Kaski, S. & Corander, J. Fundamentals and recent developments in Approximate Bayesian Computation. Syst. Biol. 66, e66–e82 (2017).
Rinta-Kokko, H., Dagan, R., Givon-Lavi, N. & Auranen, K. Estimation of vaccine efficacy against acquisition of pneumococcal carriage. Vaccine 27, 3831–3837 (2009).
Lipsitch, M. et al. Estimating rates of carriage acquisition and clearance and competitive ability for pneumococcal serotypes in Kenya with a Markov transition model. Epidemiology 23, 510–519 (2012).
Health Protection Agency COVER programme. October to December 2008. Quarterly vaccination coverage statistics for children aged up to five years in the United Kingdom. Heal. Prot. Rep. 3, 8–15 (2009).
Nuorti, J. P., Martin, S. W., Smith, P. J., Moran, J. S. & Schwartz, B. Uptake of pneumococcal conjugate vaccine among children in the 1998–2002 United States birth cohorts. Am. J. Prev. Med. 34, 46–53 (2008).
Huang, S. S., Finkelstein, J. A., Rifas-Shiman, S. L., Kleinman, K. & Platt, R. Community-level predictors of pneumococcal carriage and resistance in young children. Am. J. Epidemiol. 159, 645–654 (2004).
Durrett, R. & Levin, S. Allelopathy in spatially distributed populations. J. Theor. Biol. 185, 165–171 (1997).
Gupta, S., Ferguson, N. & Anderson, R. Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science 280, 912–915 (1998).
Henriques-Normark, B., Blomberg, C., Dagerhamn, J., Bättig, P. & Normark, S.The rise and fall of bacterial clones: Streptococcus pneumoniae. Nat. Rev. Microbiol. 6, 827–837 (2008).
Croucher, N. J. et al. Population genomic datasets describing the post-vaccine evolutionary epidemiology of Streptococcus pneumoniae. Sci. Data 2, 150058 (2015).
van Heel, A. J., de Jong, A., Montalbán-López, M., Kok, J. & Kuipers, O. P. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41, W448–W453 (2013).
Carver, T. et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–2676 (2008).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Gladman, S. VelvetOptimiser (Victorian Bioinformatics Consortium, 2010); http://www.vicbioinformatics.com/software.velvetoptimiser.shtml
Lagesen, K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 (2007).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Burge, S. W. et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 41, D226–D232 (2013).
Croucher, N. J., Vernikos, G. S., Parkhill, J. & Bentley, S. D. Identification, variation and transcription of pneumococcal repeat sequences. BMC Genomics 12, 120 (2011).
Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010).
Croucher, N. J. et al. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23F ST81. J. Bacteriol. 191, 1480–1489 (2009).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Kristensen, D. M. et al. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics 26, 1481–1487 (2010).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Page, A. J. et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2, e000056 (2016).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Cheng, L., Connor, T. R., Sirén, J., Aanensen, D. M. & Corander, J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 30, 1224–1228 (2013).
Stajich, J. E. et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12, 1611–1618 (2002).
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Croucher, N. J. et al. Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 (2011).
Pikis, A., Donkersloot, J. A., Rodriguez, W. J. & Keith, J. M. A conservative amino acid mutation in the chromosome-encoded dihydrofolate reductase confers trimethoprim resistance in Streptococcus pneumoniae. J. Infect. Dis. 178, 700–706 (1998).
Maskell, J. P., Sefton, A. M. & Hall, L. M. C. Multiple mutations modulate the function of dihydrofolate reductase in trimethoprim-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45, 1104–1108 (2001).
Haasum, Y. et al. Amino acid repetitions in the dihydropteroate synthase of Streptococcus pneumoniae lead to sulfonamide resistance with limited effects on substrate Km. Antimicrob. Agents Chemother. 45, 805–809 (2001).
Li, Y. et al. Penicillin-binding protein transpeptidase signatures for tracking and predicting β-lactam resistance levels in Streptococcus pneumoniae. mBio 7, e00756-16 (2016).
Fisher, R. A. The Genetical Theory of Natural Selection (Oxford Univ. Press, Oxford, 1930).
Wright, S. Evolution in Mendelian populations. Genetics 16, 97–159 (1931).
Der, R., Epstein, C. & Plotkin, J. B. Dynamics of neutral and selected alleles when the offspring distribution is skewed. Genetics 191, 1331–1344 (2012).
Li, Y., Thompson, C. M., Trzciński, K. & Lipsitch, M. Within-host selection is limited by an effective population of Streptococcus pneumoniae during nasopharyngeal colonization. Infect. Immun. 81, 4534–4543 (2013).
Census 2000 (US Census Bureau, 2000); https://www.census.gov/census2000/states/ma.html
Census 2011 (Office for National Statistics, 2011); http://www.ons.gov.uk/ons/guide-method/census/2011/index.html
Dutch Census 2011 (European Statistical System, 2011); https://ec.europa.eu/CensusHub2/
Turner, P. et al. A longitudinal study of Streptococcus pneumoniae carriage in a cohort of infants and their mothers on the Thailand–Myanmar border. PLoS ONE 7, e38271 (2012).
Wong, A. K. C. & You, M. Entropy and distance of random graphs with application to structural pattern recognition. IEEE Trans. Pattern Anal. Mach. Intell. 7, 599–609 (1985).
R Core Development Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2011); http://www.R-project.org/
Acknowledgements
We thank R. Gladstone, J. Jefferies, S. Faust and S. Clarke for sharing epidemiological data on the Southampton isolates. N.J.C. was funded by a Sir Henry Dale fellowship, and jointly funded by the Wellcome Trust and Royal Society (Grant Number 104169/Z/14/Z). J.C. was funded by the COIN Centre of Excellence. M.L. was funded by NIH grant R01 AI048935 and W.P.H. by NIH grant R01 AI106786.
Author information
Authors and Affiliations
Contributions
J.C., C.F., B.A., W.P.H., M.L. and N.J.C. designed the model; J.C., M.U.G. and N.J.C. fitted the model; W.P.H., S.D.B. and N.J.C. analysed the genomic data; J.C. and N.J.C. initially drafted the manuscript, with all authors contributing to the final version.
Corresponding author
Ethics declarations
Competing interests
M.L. has consulted for Pfizer, Affinivax and Merck and has received grant support not related to this paper from Pfizer and PATH Vaccine Solutions. W.P.H., M.L. and N.J.C. have consulted for Antigen Discovery Inc.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Figures 1–10; Supplementary Table 1; legends for Supplementary Datasets 1–3
Supplementary Dataset 1
Annotation of the intermediate frequency genes in the Massachusetts pneumococcal population
Supplementary Dataset 2
Annotation of the core genes in the Massachusetts pneumococcal population
Supplementary Dataset 3
Samples used in the analyses, associated epidemiological characteristics, and accession codes
Rights and permissions
About this article
Cite this article
Corander, J., Fraser, C., Gutmann, M.U. et al. Frequency-dependent selection in vaccine-associated pneumococcal population dynamics. Nat Ecol Evol 1, 1950–1960 (2017). https://doi.org/10.1038/s41559-017-0337-x
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41559-017-0337-x
This article is cited by
-
Misspecification-robust likelihood-free inference in high dimensions
Computational Statistics (2025)
-
Plasmid-driven strategies for clone success in Escherichia coli
Nature Communications (2025)
-
A genome-based survey of invasive pneumococci in Norway over four decades reveals lineage-specific responses to vaccination
Genome Medicine (2024)
-
Genomic and panproteomic analysis of the development of infant immune responses to antigenically-diverse pneumococci
Nature Communications (2024)
-
Mathematical Modelling of Parasite Dynamics: A Stochastic Simulation-Based Approach and Parameter Estimation via Modified Sequential-Type Approximate Bayesian Computation
Bulletin of Mathematical Biology (2024)