Abstract
More than one-third of the world’s population is exposed to Plasmodium vivax malaria, mainly in Asia1. P. vivax preferentially invades reticulocytes (immature red blood cells)2,3,4. Previous work has identified 11 parasite proteins involved in reticulocyte invasion, including erythrocyte binding protein 2 (ref. 5) and the reticulocyte-binding proteins (PvRBPs)6,7,8,9,10. PvRBP2b binds to the transferrin receptor CD71 (ref. 11), which is selectively expressed on immature reticulocytes12. Here, we identified CD98 heavy chain (CD98), a heteromeric amino acid transporter from the SLC3 family (also known as SLCA2), as a reticulocyte-specific receptor for the PvRBP2a parasite ligand using mass spectrometry, flow cytometry, biochemical and parasite invasion assays. We characterized the expression level of CD98 at the surface of immature reticulocytes (CD71+) and identified an interaction between CD98 and PvRBP2a expressed at the merozoite surface. Our results identify CD98 as an additional host membrane protein, besides CD71, that is directly associated with P. vivax reticulocyte tropism. These findings highlight the potential of using PvRBP2a as a vaccine target against P. vivax malaria.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Source data are provided with this paper. All other data are available from the corresponding authors upon reasonable request.
References
Confronting Plasmodium vivax malaria. WHO/HTM/GMP/2015.3 (World Health Orgainization, 2015).
Hegner, R. Relative frequency of ring-stage plasmodia in reticulocytes and mature erythrocytes in man an monkey. Am. J. Trop. Med. Hyg. 27, 690–718 (1938).
Mons, B., Croon, J. J., van der Star, W. & van der Kaay, H. J. Erythrocytic schizogony and invasion of Plasmodium vivax in vitro. Int. J. Parasitol. 18, 307–311 (1988).
Malleret, B. et al. Plasmodium vivax: restricted tropism and rapid remodeling of CD71-positive reticulocytes. Blood 125, 1314–1324 (2015).
Hester, J. et al. De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Negl. Trop. Dis. 7, e2569 (2013).
Carlton, J. M. et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455, 757–763 (2008).
Han, J. H. et al. Identification of a reticulocyte-specific binding domain of Plasmodium vivax reticulocyte-binding protein 1 that is homologous to the PfRh4 erythrocyte-binding domain. Sci. Rep. 6, 26993 (2016).
Gupta, S. et al. Targeting a reticulocyte binding protein and Duffy binding protein to inhibit reticulocyte invasion by Plasmodium vivax. Sci. Rep. 8, 10511 (2018).
Ntumngia, F. B. et al. Identification and immunological characterization of the ligand domain of Plasmodium vivax reticulocyte binding protein 1a. J. Infect. Dis. 218, 1110–1118 (2018).
Chim-Ong, A. et al. The blood stage antigen RBP2-P1 of Plasmodium vivax binds reticulocytes and is a target of naturally acquired immunity. Infect. Immun. 88, e00616–e00619 (2020).
Gruszczyk, J. et al. Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science 359, 48–55 (2018).
Malleret, B. et al. Significant biochemical, biophysical and metabolic diversity in circulating human cord blood reticulocytes. PLoS ONE 8, e76062 (2013).
Proto, W. R. et al. Adaptation of Plasmodium falciparum to humans involved the loss of an ape-specific erythrocyte invasion ligand. Nat. Commun. 10, 4512 (2019).
Kosaisavee, V. et al. Strict tropism for CD71+/CD234+ human reticulocytes limits the zoonotic potential of Plasmodium cynomolgi. Blood 130, 1357–1363 (2017).
Wright, G. J. & Rayner, J. C. Plasmodium falciparum erythrocyte invasion: combining function with immune evasion. PLoS Pathog. 10, e1003943 (2014).
Crosnier, C. et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature 480, 534–537 (2011).
Douglas, A. D. et al. Neutralization of Plasmodium falciparum merozoites by antibodies against PfRH5. J. Immunol. 192, 245–258 (2014).
Barnwell, J. W., Nichols, M. E. & Rubinstein, P. In vitro evaluation of the role of the Duffy blood group in erythrocyte invasion by Plasmodium vivax. J. Exp. Med. 169, 1795–1802 (1989).
Malleret, B., Renia, L. & Russell, B. The unhealthy attraction of Plasmodium vivax to reticulocytes expressing transferrin receptor 1 (CD71). Int. J. Parasitol. 47, 379–383 (2017).
Kanjee, U. et al. Plasmodium vivax strains use alternative pathways for invasion. J. Infect. Dis. 223, 1817–1821 (2020).
Chu, T. T. T. et al. Quantitative mass spectrometry of human reticulocytes reveal proteome-wide modifications during maturation. Br. J. Haematol. 180, 118–133 (2018).
Boado, R. J., Li, J. Y., Nagaya, M., Zhang, C. & Pardridge, W. M. Selective expression of the large neutral amino acid transporter at the blood–brain barrier. Proc. Natl Acad. Sci. USA 96, 12079–12084 (1999).
Segawa, H. et al. Identification and functional characterization of a Na+-independent neutral amino acid transporter with broad substrate selectivity. J. Biol. Chem. 274, 19745–19751 (1999).
Fort, J. et al. The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane. J. Biol. Chem. 282, 31444–31452 (2007).
Yan, R., Zhao, X., Lei, J. & Zhou, Q. Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature 568, 127–130 (2019).
Chiduza, G. N. et al. LAT1 (SLC7A5) and CD98hc (SLC3A2) complex dynamics revealed by single-particle cryo-EM. Acta Crystallogr. D Struct. Biol. 75, 660–669 (2019).
Lee, Y. et al. Cryo-EM structure of the human l-type amino acid transporter 1 in complex with glycoprotein CD98hc. Nat. Struct. Mol. Biol. 26, 510–517 (2019).
Russell, B. et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood 118, e74–e81 (2011).
Friguet, B., Chaffotte, A. F., Djavadi-Ohaniance, L. & Goldberg, M. E. Measurements of the true affinity constant in solution of antigen–antibody complexes by enzyme-linked immunosorbent assay. J. Immunol. Methods 77, 305–319 (1985).
Batchelor, J. D. et al. Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS Pathog. 10, e1003869 (2014).
Tham, W. H. et al. Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand. Proc. Natl Acad. Sci. USA 107, 17327–17332 (2010).
Srivastava, A. et al. Host reticulocytes provide metabolic reservoirs that can be exploited by malaria parasites. PLoS Pathog. 11, e1004882 (2015).
Obaldia, N. III et al. Bone marrow is a major parasite reservoir in Plasmodium vivax infection. mBio 9, e00625-18 (2018).
Galinski, M. R., Medina, C. C., Ingravallo, P. & Barnwell, J. W. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell 69, 1213–1226 (1992).
Gruszczyk, J. et al. Structurally conserved erythrocyte-binding domain in Plasmodium provides a versatile scaffold for alternate receptor engagement. Proc. Natl Acad. Sci. USA 113, E191–E200 (2016).
Franca, C. T. et al. Plasmodium vivax reticulocyte binding proteins are key targets of naturally acquired immunity in young Papua New Guinean children. PLoS Negl. Trop. Dis. 10, e0005014 (2016).
Ryan, J. R. et al. Evidence for transmission of Plasmodium vivax among a duffy antigen negative population in Western Kenya. Am. J. Trop. Med. Hyg. 75, 575–581 (2006).
Menard, D. et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc. Natl Acad. Sci. USA 107, 5967–5971 (2010).
Cavasini, C. E. et al. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans. R. Soc. Trop. Med. Hyg. 101, 1042–1044 (2007).
Chitnis, C. E. & Miller, L. H. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180, 497–506 (1994).
Keller, A., Nesvizhskii, A. I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Nesvizhskii, A. I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Sriprawat, K. et al. Effective and cheap removal of leukocytes and platelets from Plasmodium vivax infected blood. Malar. J. 8, 115 (2009).
Wasniowska, K. et al. Structural characterization of the epitope recognized by the new anti-Fy6 monoclonal antibody NaM 185-2C3. Transfus. Med 12, 205–211 (2002).
Lee, W. C. et al. Glycophorin C (CD236R) mediates vivax malaria parasite rosetting to normocytes. Blood 123, e100–e109 (2014).
Peng, K. et al. Breadth of humoral response and antigenic targets of sporozoite-inhibitory antibodies associated with sterile protection induced by controlled human malaria infection. Cell Microbiol. 18, 1739–1750 (2016).
Carissimo, G. et al. VCP/p97 Is a proviral host factor for replication of chikungunya virus and other alphaviruses. Front. Microbiol. 10, 2236 (2019).
Acknowledgements
We are indebted to C. Chu, R. McGready and the staff of the Mae Sot Malaria Clinic and the clinics associated with SMRU (Tak Province, Thailand) and the patients attending these clinics. We thank M. Mauduit for help at the beginning of the project. We thank the SIgN flow cytometry platform (supported by a grant from the National Research Foundation, Immunomonitoring Service Platform ISP) (NRF2017_SISFP09)). B.R. and B.M. were funded by the Singapore National Medical Research Council (NMRC/CBRG/0047/2013). B.M. was also funded by the Agency for Science, Technology and Research (A*STAR, Singapore) Young Investigator Grant (BMRC YIG grant no: 13/1/16/YA/009), core funds to SIgN from A*STAR, NUHS Start-up grant (NUHSRO/2018/006/SU/01) and MOE Tier 1 (NUHSRO/2018/094/T1/SEED-NOV/04). L.R. was supported by a Singapore National Medical Research Council IRG grant (NMRC/OFIRG/0065/2018), by a Singapore Immunology Network core research grant and by the Horizontal Programme on Infectious Diseases under A*STAR. SMRU is supported by The Wellcome Trust of Great Britain as part of the Oxford Tropical Medicine Research Programme of Wellcome Trust–Mahidol University. R.C. acknowledges funding support through the following grants: T1MOE1702 (MOE Tier 1 Grant through the Singapore University of Technology & Design) and RGUOO180301 (Marsden Grant Sub-award through the University of Otago). W.-H.T. was funded by the Australian Research Council Future Fellowship. Duke-NUS Medical School efforts were supported by Singapore’s Health and Biomedical Sciences (HBMS) Industry Alignment Fund Pre-Positioning (IAF-PP) grant H18/01/a0/018, administered by A*STAR.
Author information
Authors and Affiliations
Contributions
B.M. and B.R. carried out the phenotyping characterization of the erythrocytes and the antibody validations for the P. vivax invasion assays. G.C., S.W.H., R.S., V.K. and A.S.M.O. developed the P. vivax library and performed the erythrocyte binding assays. T.T.T.C., A.S. and R.C. performed and analysed the MS data. J.G. and W.-H.T. carried out construct design and protein purification for the PvRBP2a recombinant proteins and the anti-PvRBP2a monoclonal antibodies. Y.C. developed the anti-Duffy antibodies. J.K.Y.C., Y.F. and F.N. were in charge of the management of clinical data. A.E.S., J. Lin, J. Lescar, G.C. and L.F.P.N. managed the biochemistry aspects of the project. S.M.-S. managed the protein modelling of the interaction between CD98 and PvRPB2a. W.N., M.Z.T. and A.-M.C. managed the Octet experiments for CD98 and PvRPB2a interaction measurements. Overall project management was carried out by B.M., B.R. and L.R. The manuscript was prepared by B.M., G.S., B.R. and L.R.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Microbiology thanks Tuan Tran, Sanjeeva Srivastava and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
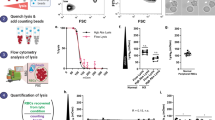
Extended Data Fig. 1 Reticulocyte phenotyping.
Trypsin resistance profile of different markers expressed at the surface of cord blood reticulocytes. The black and yellow histograms represent the level of expression before and after trypsin treatment respectively. The trypsin sensitive proteins are annotated in red and the resistant ones in black. The isotype antibody used as control is represented in grey. We used human cord blood samples pre-enriched with reticulocytes and that thus still harbour some normocytes, hence the double peak observed for some proteins such as CD71 and CD98.
Extended Data Fig. 2 Flow cytometry analysis of adult peripheral blood, cord blood and P. vivax isolates.
a, Flow cytometry profile of CD98 and CD71 expression on thiazole orange (TO) negative, low and high erythrocytes for three different human adult peripheral blood samples (forward scatter (FSC) for x-axis and side scatter (SSC) for y-axis). The TO high subset represents the most immature reticulocyte population compared to low subset. b, Flow cytometry histogram of CD98 expression (right) at the surface of P. vivax rings, early infected reticulocyte forms, gated on Hoechst-positive cells (left), side scatter (SSC) for y-axis. The infected cells positive for Hoechst (a DNA stain) are in blue and the uninfected red blood cells (uRBC), which do not contain DNA, are in red. c, Comparison of delta of geometric mean fluorescence intensity (MFI) between immature reticulocytes from three cord blood samples and three vivax infected patients (ring stage), Values are expressed as mean ± SD, unpaired Student t-test.
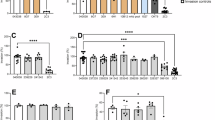
Extended Data Fig. 3 Binding assay of erythrocytes to HEK cells expressing P. vivax genes.
a, Schematic description of the development of the library and its use in the erythrocyte binding assay: (1) cloning of P. vivax genes in the pDisplay plasmid, (2) transfection of HEK cells, (3) expression of the protein containing MYC and HA tags at the surface of HEK cells and (5) binding assay with erythrocytes. b, Schematic representation of full-length PvRBP2a and of its recombinant protein fragments used in this study (right). Signal peptide (SP), transmembrane helix (TM) and nucleotide-binding domain (green) are indicated. The ticks indicated for nonsynonymous SNPs with alternate allele frequency >20%. c, Representative images from 3 independent experiments of binding of reticulocytes (CD71 + ) or normocytes (CD71-) loaded with the fluorescent dye CFSE to HEK cells expressing the PvRBP2a 1315-1650 (negative control), Duffy binding protein II (DBPII) fragment or PvRBP2a23-767 in the presence or absence of anti-CD98 antibodies. The scale represents 50 μm. d, The binding of reticulocytes to HEK transfected with the PvRBP2a23–767 fragment was done in octuplicate (eight independent experiments) and was shown to follow a normal distribution as determine by the D’Agostino’s K-squared test, and differed significantly from binding to non-transfected HEK cells, Values are expressed as mean ± SD, Student t-test. e, Binding assays of reticulocytes to HEK cells expressing PvRBP2b481-1530 in the presence or absence of anti-CD98 antibodies. (three independent experiments). Values are expressed as mean ± SD, no significant differences were observed. e, Binding assays of normocytes to HEK cells expressing PvRBP2a23-767 in the presence or absence of anti-CD98 antibodies (three independent experiments). Values are expressed as mean ± SD, no significant differences were observed.
Extended Data Fig. 4 Antigen specificity of anti-PvRBP2a antibodies.
Mouse 1C3 and 3A11 mAbs recognize specifically the PvRBP2a fragment, PvRBP2a23–767, expressed by HEK cells when tested by flow cytometry. As positive control, rabbit anti-myc antibodies were used. Secondary anti-mouse IgG antibodies coupled with eFluor 660 or mouse anti-rabbit immunoglobulin coupled with Alexa 674 were used as negative control.
Supplementary information
Supplementary Tables
Combined PDF with Supplementary Tables 1–5.
Source data
Source Data Fig. 1
Raw data.
Source Data Fig. 1
Unprocessed western blot.
Source Data Fig. 2
Raw data.
Source Data Fig. 3
Raw data.
Source Data Fig. 4
Raw data.
Source Data Fig. 4
Unprocessed immunoprecipitation gel.
Source Data Extended Data Fig. 2
Raw data.
Source Data Extended Data Fig. 3
Raw data.
Rights and permissions
About this article
Cite this article
Malleret, B., El Sahili, A., Tay, M.Z. et al. Plasmodium vivax binds host CD98hc (SLC3A2) to enter immature red blood cells. Nat Microbiol 6, 991–999 (2021). https://doi.org/10.1038/s41564-021-00939-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-021-00939-3
This article is cited by
-
The pattern of gene amplification of members of the Plasmodium vivax erythrocyte binding-like proteins family across the Amazon rainforest
Malaria Journal (2025)
-
Highly conserved Plasmodium vivax genomes in Duffy-negative individuals from Sudan
Scientific Reports (2025)
-
Genomic analysis of Plasmodium vivax field isolates circulating in sub-Saharan Africa
Communications Biology (2025)
-
Intestinal Goblet Cell-Expressed Reg4 Ameliorates Intestinal Inflammation Potentially by Restraining Pathogenic Escherichia coli Infection
Probiotics and Antimicrobial Proteins (2025)
-
The PvRBP2b-TfR1 interaction is not essential for reticulocytes invasion by Plasmodium vivax isolates from Cambodia
npj Vaccines (2024)