Abstract
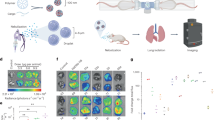
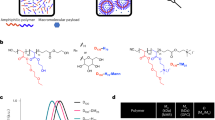
Here we describe formulations comprising individual, polymer-complexed self-amplifying RNA (saRNA) molecules, designed for vaccination against infectious diseases and other prophylactic and therapeutic applications. When exposed to a large excess of the cationic polymer polyethylenimine (PEI), the single saRNA molecules in solution reorganize from an extended to a globular organization, characterized by a high packing density, low polymer mass fraction and, consequently, a very small size of the polyplex nanoparticles of about 30 nm. This format of PEI-complexed saRNA exhibits enhanced biological activity in comparison with previously described saRNA/PEI formulations, both in vitro and in vivo. In vaccination models, relevant immune responses at lower doses are achieved, offering potential advantages for practical use. We found that the single PEI-complexed RNA molecules are also present in conventional formulations to some degree. The direct correlation between the single-molecule fraction with activity suggests that it is this format that predominantly contributes to activity in the different formulation types. Complexation is driven by mechanisms of self-assembly between oppositely charged polyelectrolytes, making this protocol broadly applicable to various cationic polymers and RNA constructs. With their small size and good stability in biofluids, these compacted RNA molecules are also promising for the systemic delivery of genetic material to compartments that are difficult to reach with larger particles.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request. For the SANS measurements, the raw images are public via ISIS Neutron Source at https://data.isis.stfc.ac.uk/doi/INVESTIGATION/111243135/ and https://doi.org/10.5286/ISIS.E.RB2000156. Source data are provided with this paper.
References
Labouta, H. I. et al. Role of drug delivery technologies in the success of COVID-19 vaccines: a perspective. Drug Deliv. Transl. Res. 12, 2581–2588 (2022).
Sahin, U., Karikó, K. & Türeci, Ö. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014).
Chaudhary, N., Weissman, D. & Whitehead, K. A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov. 20, 880 (2021).
Bloom, K., van den Berg, F. & Arbuthnot, P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 28,117–129 (2020).
Vogel, A. B. et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 26, 446–455 (2018).
Pollock, K. M. et al. Safety and immunogenicity of a self-amplifying RNA vaccine against COVID-19: COVAC1, a phase I, dose-ranging trial. EClinicalMedicine 44, 101262 (2022).
Davis, N. L., Willis, L. V., Smitht, J. F. & Johnston, R. E. In vitro synthesis of infectious venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology 171, 189–204 (1989).
Tenchov, R. et al. Lipid nanoparticles: from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano 15, 16982–17015 (2021).
Samaridou, E., Heyes, J. & Lutwyche, P. Lipid nanoparticles for nucleic acid delivery: current perspectives. Adv. Drug Deliv. Rev. 154–155, 37–63 (2020).
Kranz, L. M. et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016).
Aldon, Y. et al. Immunogenicity of stabilized HIV-1 Env trimers delivered by self-amplifying mRNA. Mol. Ther. Nucleic Acids 25, 483–493 (2021).
Blakney, A. K. et al. Polymeric and lipid nanoparticles for delivery of self-amplifying RNA vaccines. J. Control. Release 338, 201–210 (2021).
Blakney, A. K. et al. Big is beautiful: enhanced saRNA delivery and immunogenicity by a higher molecular weight, bioreducible, cationic polymer. ACS Nano 14, 5711–5727 (2020).
Démoulins, T. et al. Polyethylenimine-based polyplex delivery of self-replicating RNA vaccines. Nanomed. Nanotechnol. Biol. Med. 12, 711–722 (2016).
Casper, Jens et al. Polyethylenimine (PEI) in gene therapy: current status and clinical applications. J. Control. Release 362, 667–691 (2023).
Boussif, O. et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA 92, 7297–7301 (1995).
Zou, S. M., Erbacher, P., Remy, J. S. & Behr, J. P. Systemic linear polyethylenimine (L‐PEI)‐mediated gene delivery in the mouse. J. Gene Med. 2, 128–134 (2000).
Bonnet, M.-E., Erbacher, P. & Bolcato-Bellemin, A.-L. Systemic delivery of DNA or siRNA mediated by linear polyethylenimine (L-PEI) does not induce an inflammatory response. Pharm. Res. 25, 2972–2982 (2008).
Perevyazko, I. Y. et al. Polyelectrolyte complexes of DNA and linear PEI: formation, composition and properties. Langmuir 28, 16167–16176 (2012).
Gallops, C. E., Yu, C., Ziebarth, J. D. & Wang, Y. Effect of the protonation level and ionic strength on the structure of linear polyethyleneimine. ACS Omega 4, 7255–7264 (2019).
Huh, S.-H. et al. Optimization of 25 kDa linear polyethylenimine for efficient gene delivery. Biologicals 35, 165–171 (2007).
Parot, J., Caputo, F., Mehn, D., Hackley, V. A. & Calzolai, L. Physical characterization of liposomal drug formulations using multi-detector asymmetrical-flow field flow fractionation. J. Control. Release 320, 495–510 (2020).
Graewert, M. A. et al. Quantitative size-resolved characterization of mRNA nanoparticles by in-line coupling of asymmetrical-flow field-flow fractionation with small angle X-ray scattering. Sci. Rep. 13, 15764 (2023).
Guerrini, G. et al. Analytical ultracentrifugation to assess the quality of LNP–mRNA therapeutics. Int. J. Mol. Sci. 25, 5718 (2024).
Oliver, R. C., Rolband, L. A., Hutchinson-Lundy, A. M., Afonin, K. A. & Krueger, J. K. Small-angle scattering as a structural probe for nucleic acid nanoparticles (NANPs) in a dynamic solution environment. Nanomaterials 9, 681 (2019).
Takamoto, K. et al. Principles of RNA compaction: insights from the equilibrium folding pathway of the P4–P6 RNA domain in monovalent cations. J. Mol. Biol. 343, 1195–1206 (2004).
Guzmán-Terán, C., Calderón-Rangel, A., Rodriguez-Morales, A. & Mattar, S. Venezuelan equine encephalitis virus: the problem is not over for tropical America. Ann. Clin. Micro. Antimicrob 19, 19 (2020).
Abhijeet, D. et al. Polyelectrolyte complexes: mechanisms, critical experimental aspects, and applications. Artif. Cells Nanomed. Biotechnol. 44, 1615–1625 (2016).
Haas, H., Moreno Herrero, J., Schlegel, A. M. G. & Erbar, S. RNA formulations suitable for therapy. US patent WO2021001417A1 (2021).
Moreno Herrero, J., Haas, H., Erbar, S. & Stahl, T. B. Nucleic acid complexes and uses thereorf. US patent WO2024068674A1 (2024).
Siewert, C. et al. Investigation of charge ratio variation in mRNA—DEAE–dextran polyplex delivery systems. Biomaterials 192, 612–620 (2019).
Manalastas-Cantos, K. et al. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 54, 343–355 (2021).
Kuhn, A. N. et al. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 17, 961–971 (2010).
Pokrovskaya, I. D. & Gurevich, V. V. In vitro transcription: preparative RNA yields in analytical scale reactions. Anal. Biochem. 220, 420–423 (1994).
Haas, H., Erbar, S. & Heidenreich, R. Formulation for administration of RNA. US patent WO2018011406A1 (2018).
Erbacher, P. et al. Genuine DNA/polyethylenimine (PEI) complexes improve transfection properties and cell survival. J. Drug Target. 12, 223–236 (2008).
Vita, R. et al. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343 (2019).
Wang, P. et al. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 4, e1000048 (2008).
Paul, S. et al. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J. Immunol. 191, 5731–5739 (2013).
Blanchet, C. E. et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 48, 431–443 (2015).
Schroer, M. A. et al. Smaller capillaries improve the small-angle X-ray scattering signal and sample consumption for biomacromolecular solutions. J. Synchrotron Radiat. 25, 1113–1122 (2018).
Round, A. et al. BioSAXS sample changer: a robotic sample changer for rapid and reliable high-throughput X-ray solution scattering experiments. Acta Crystallogr. D Biol. Crystallogr. 71, 67–75 (2015).
Franke, D., Kikhney, A. G. & Svergun, D. I. Automated acquisition and analysis of small angle X-ray scattering data. Nucl. Instrum. Methods Phys. Res. A 689, 52–59 (2012).
Svergun, D. I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992).
Franke, D. & Svergun, D. I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009).
Arnold, O. et al. Mantid—data analysis and visualization package for neutron scattering and μ SR experiments. Nucl. Instrum. Methods Phys. Res. A 764, 156–166 (2014).
Rubinson, K. A., Stanley, C. & Krueger, S. Small-angle neutron scattering and the errors in protein structures that arise from uncorrected background and intermolecular interactions. J. Appl. Crystallogr. 41, 456–465 (2008).
Orthaber, D., Bergman, A. & Glatter, O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Crystallogr. 33, 218–225 (2000).
Ibel, K. & Stuhrmann, H. B. Comparison of neutron and X-ray scattering of dilute myoglobin solutions. J. Mol. Biol. 93, 255–265 (1975).
Stuhrmann, H. B. Small-angle scattering and its interplay with crystallography, contrast variation in SAXS and SANS. Acta Crystallogr. A A64, 181–191 (2008).
Acknowledgements
Figures 1, 3 and 4 were illustrated and designed by Elvire Thouvenot and are reproduced with permission, copyright 2022 Elvire Thouvenot. We thank JASCO and S. Suzuki for making available the HTS Module for early CD measurements and helping throughout the establishment of the HTS protocol used in this Article. We acknowledge A. Westenberger for excellent technical assistance. For SANS data analysis, this work benefitted from the use of the SasView application, originally developed under NSF award DMR-0520547. For SANS data analysis, the software package SasView was used. SasView contains code developed with funding from the European Union’s Horizon 2020 research and innovation programme under the SINE2020 project, grant agreement no. 654000.
Author information
Authors and Affiliations
Contributions
Conceptualization by J.M.H., S.E., A.S. and H.H. Methodology by J.M.H., S.E., J.S., A.S. and H.H. Investigation by J.M.H., T.B.S., J.S., K.M., T.B., L.P.C., M.A.S. and D.I.S. Visualization by J.M.H., S.E. and H.H. Project administration by S.E., A.S., H.H. and U.S. Supervision by S.E., A.S., H.H. and U.S. Writing—original draft by J.M.H., S.E. and H.H. Writing— review and editing by J.M.H., T.B.S., S.E., K.M., A.S., J.S., T.B., L.P.C., M.A.S., D.I.S. and H.H.
Corresponding authors
Ethics declarations
Competing interests
U.S. is a management board member at BioNTech SE. T.B.S., J.S. and T.B. are employees at BioNTech SE. J.M.H., T.B.S., S.E., A.S., T.B., H.H. and U.S. are inventors on patents and patent applications related to RNA technology and/or have securities from BioNTech SE. All other authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks Gero Decher, Reidar Lund and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary data, Figs. 1–8 and Tables 1 and 2.
Source data
Source data Fig. 1
Excel file with data for Fig. 1.
Source data Fig. 2C
Excel file with data for Fig. 2c.
Source data Fig. 4
Excel file with data for Fig. 4.
Source data Fig. 5
Excel file with data for Fig. 5.
Source data Fig. 6
Excel file with data for Fig. 6.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Moreno Herrero, J., Stahl, T.B., Erbar, S. et al. Compact polyethylenimine-complexed mRNA vaccines. Nat. Nanotechnol. 20, 1323–1331 (2025). https://doi.org/10.1038/s41565-025-01961-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41565-025-01961-w