Abstract
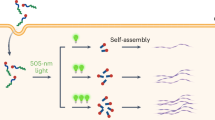
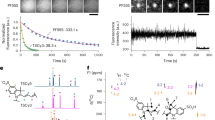
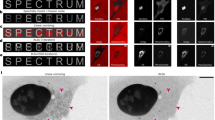
Fluorescent nanoparticles offer superior brightness and photostability compared with conventional dyes and proteins. However, their relatively large size and complex surface chemistry limit their utility for imaging nanoscale biostructures and tracking individual proteins in living cells. Here we develop single-chain ultrasmall fluorescent polymer dots (suPdots) with size below 5 nm, comparable to fluorescent proteins. Fabricated via vitrification of conjugated polymer solutions, suPdots enable tunable fluorescence as well as high-density, specific labelling of multiple subcellular organelles. We demonstrate nanoscopic imaging of continuous ring structures in clathrin-coated pits as well as multi-target stimulated emission depletion imaging. Thanks to their high brightness, suPdots enable tracking the individual steps of the kinesin-1 motor protein in living cells using standard spinning-disk fluorescence microscopy, with a 16-nm step size and 50-Hz temporal resolution. These demonstrations establish suPdots as powerful, versatile fluorescent probes for nanoscale-resolution biomolecular imaging with increased accessibility and efficiency for diverse bio-applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon request. Source data are provided with this paper.
References
Francisco, B. et al. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355, 606–612 (2016).
Li, D. et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349, aab3500 (2015).
Ouyang, Z. et al. Elucidating subcellular architecture and dynamics at isotropic 100-nm resolution with 4Pi-SIM. Nat. Methods 22, 335–347 (2024).
Nehme, E. et al. DeepSTORM3D: dense 3D localization microscopy and PSF design by deep learning. Nat. Methods 17, 734–740 (2020).
Huang, X. et al. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat. Biotechnol.36, 451–459 (2018).
Gwosch, K. C. et al. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods 17, 217–224 (2020).
Day, R. N. & Davidson, M. W. The fluorescent protein palette: tools for cellular imaging. Chem. Soc. Rev.38, 2887–2921 (2009).
Zeng, S. et al. Fluorescent dyes based on rhodamine derivatives for bioimaging and therapeutics: recent progress, challenges, and prospects. Chem. Soc. Rev.52, 5607–5651 (2023).
Jin, D. et al. Nanoparticles for super-resolution microscopy and single-molecule tracking. Nat. Methods 15, 415–423 (2018).
Ashoka, A. H. et al. Brightness of fluorescent organic nanomaterials. Chem. Soc. Rev.52, 4525–4548 (2023).
Xu, Y. et al. Recent advances in luminescent materials for super-resolution imaging via stimulated emission depletion nanoscopy. Chem. Soc. Rev. 50, 667–690 (2021).
Wegner, K. D. et al. Quantum dots: bright and versatile in vitro and in vivo fluorescence imaging biosensors. Chem. Soc. Rev. 44, 4792–4834 (2015).
Li, W. et al. Fluorescent nanoparticles for super-resolution imaging. Chem. Rev. 122, 12495–12543 (2022).
Hoffman, D. P. et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367, eaaz5357 (2020).
Silvester, E., et al. DNA origami signposts for identifying proteins on cell membranes by electron cryotomography. Cell 184, 1110–1121 (2021).
Tao, H., et al. Nanoparticle synthesis assisted by machine learning. Nat. Rev. Mater. 6, 701–716 (2021).
Li, K., et al. Polymer-encapsulated organic nanoparticles for fluorescence and photoacoustic imaging. Chem. Soc. Rev. 43, 6570–6597 (2014).
Takahiro, D., et al. Direct observation of motor protein stepping in living cells using MINFLUX. Science 379, 1010–1015 (2023).
Jan, O. W., et al. MINFLUX dissects the unimpeded walking of kinesin-1. Science 379, 1004–1010 (2023).
Fan, Q., et al. Precise control over kinetics of molecular assembly: production of particles with tunable sizes and crystalline forms. Angew. Chem. Int. Ed. 59, 15141–15146 (2020).
Yan, Z., et al. Preparation of ultrasmall AIE nanoparticles with tunable molecular packing via freeze assembly. Nano Lett. 23, 1030–1035 (2023).
Mao, J., et al. Templated freezing assembly precisely regulates molecular assembly for free-standing centimeter-scale microtextured nanofilms. Sci. China Chem. 66, 878–886 (2023).
Carrie, L., et al. Effects of packing structure on the optoelectronic and charge transport properties in poly(9-9-di-n-octylfluorene-alt-benzothiadiazole). J. Am. Chem. Soc. 127, 12890–12899 (2005).
Wang, D., et al. From single chains to aggregates, how conjugated polymers behave in dilute solutions. Macromolecules 46, 6217–6224 (2013).
Wu, C., et al. Highly fluorescent semiconducting polymer dots for biology and medicine. Angew. Chem. Int. Ed. 52, 3086–3109 (2013).
Osamu Mishima, H. E. S. The relationship between liquid, supercooled and glassy water. Nature 396, 329–335 (1998).
Guan, W., et al. Measurement of solubilization location in micelles using anchored aggregation-induced emission donors. Angew. Chem. Int. Ed. 59, 12800–12805 (2020).
Zhang, J., et al. Ultrabright Pdots with a large absorbance cross section and high quantum yield. ACS Appl. Mater. Interfaces 14, 13631–13637 (2022).
Wang, S., et al. NIR-II excitable conjugated polymer dots with bright NIR-I emission for deep in vivo two-photon brain imaging through intact skull. Adv. Funct. Mater. 29, 1808365 (2019).
Chen, Y., et al. Surface modification of gold nanoparticles with small molecules for biochemical analysis. Acc. Chem. Res. 50, 310–319 (2017).
Gajdos, T., et al. Hot-band anti-Stokes fluorescence properties of Alexa Fluor 568. J. Fluoresc. 30, 437–443 (2020).
Xu, J., et al. Automated stoichiometry analysis of single-molecule fluorescence imaging traces via deep learning. J. Am. Chem. Soc. 141, 6976–6985 (2019).
Tang, D., et al. Topological DNA tetrahedron encapsulated gold nanoparticle enables precise ligand engineering for targeted cell imaging. Anal. Chem. 93, 17036–17042 (2021).
Paul, F. B., et al. Hole-induced quenching of triplet and singlet excitons in conjugated polymers. J. Am. Chem. Soc. 127, 9556–9560 (2005).
Paul, F. B., et al. Discrete intensity jumps and intramolecular electronic energy transfer in the spectroscopy of single conjugated polymer molecules. Science 277, 1074–1077 (1997).
Paul, F. B. et al. Size-dependent spectroscopic properties of conjugated polymer nanoparticles. J. Phys. Chem. B 110, 25568–25572 (2006).
Chen, X. et al. Small photoblinking semiconductor polymer dots for fluorescence nanoscopy. Adv. Mater. 29, 1604850 (2017).
Chen, X. et al. Multicolor super-resolution fluorescence microscopy with blue and carmine small photoblinking polymer dots. ACS Nano 11, 8084–8091 (2017).
Paul, F. B. et al. Collapse of stiff conjugated polymers with chemical defects into ordered, cylindrical conformations. Nature 405, 1030–1033 (2000).
Joshua, C. B. et al. Conformation and energy transfer in single conjugated polymers. Acc. Chem. Res. 45, 1992–2001 (2012).
Jiang, Y., et al. Light-harvesting and amplified energy transfer in conjugated polymer nanoparticles. Chem. Rev. 117, 838–859 (2017).
Xu, X. et al. Influence of side-chain length and relative rigidities of backbone and side chains on glass formation of branched polymers. Macromolecules 54, 6327–6341 (2021).
Wu, C. et al. Bioconjugation of ultrabright semiconducting polymer dots for specific cellular targeting. J. Am. Chem. Soc.132, 15410–15417 (2010).
Wu, Y. et al. Fluorescent polymer dot-based multicolor stimulated emission depletion nanoscopy with a single laser beam pair for cellular tracking. Anal. Chem. 92, 12088–12096 (2020).
Yu, J., et al. Recent advances in the development of highly luminescent semiconducting polymer dots and nanoparticles for biological imaging and medicine. Anal. Chem. 89, 42–56 (2016).
Rittweger, E., et al. STED microscopy reveals crystal colour centres with nanometric resolution. Nat. Photon. 3, 144–147 (2009).
Bates, M. et al. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007).
Anderson, J., et al. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J. Cell Biol. 108, 389–400 (1989).
Liang, L., et al. Continuous-wave near-infrared stimulated-emission depletion microscopy using downshifting lanthanide nanoparticles. Nat. Nanotechnol. 16, 975–980 (2021).
Zhan, Q., et al. Achieving high-efficiency emission depletion nanoscopy by employing cross relaxation in upconversion nanoparticles. Nat. Commun. 8, 1058 (2017).
Hanne, J. et al. STED nanoscopy with fluorescent quantum dots. Nat. Commun.6, 7127 (2015).
Pisfil, M. G. et al. Stimulated emission depletion microscopy with a single depletion laser using five fluorochromes and fluorescence lifetime phasor separation. Sci. Rep.12, 14027–14042 (2022).
Blom, H. et al. Stimulated emission depletion microscopy. Chem. Rev. 117, 7377–7427 (2017).
Peng, C. S. et al. Nanometer-resolution tracking of single cargo reveals dynein motor mechanisms. Nat. Chem. Biol. 21, 648–656 (2024).
Ahmet, Y. et al. Kinesin walks hand-over-hand. Science 303, 676–678 (2004).
Niekamp, S. et al. A 6-nm ultra-photostable DNA FluoroCube for fluorescence imaging. Nat. Methods17, 437–441 (2020).
Hiroyasu, H., et al. Live-cell single-molecule labeling and analysis of myosin motors with quantum dots. Mol. Biol. Cell 28, 173–181 (2016).
Sun, C. et al. Intracellular tracking of single native molecules with electroporation-delivered quantum dots. Anal. Chem. 86, 11403–11409 (2014).
Wichner, S. M., et al. Covalent protein labeling and improved single-molecule optical properties of aqueous CdSe/CdS quantum dots. ACS Nano 11, 6773–6781 (2017).
Tinevez, J.-Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Loeff, L., et al. AutoStepfinder: a fast and automated step detection method for single-molecule analysis. Patterns 2, 100256 (2021).
Steffen, J. S. et al. Direct optical measurement of intramolecular distances with angstrom precision. Science 386, 180–187 (2024).
Zhang, X., et al. Importance of having low-density functional groups for generating high-performance semiconducting polymer dots. ACS Nano 6, 5429–5439 (2012).
Ershov, D., et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 19, 829–932 (2022).
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (grant numbers T2293760 and T2293762 to J.W. and 52573355 to Q.F.), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant numbers XDB1030000 to J.W. and Q.F., and XDB1020000 to X.F.) and the National Key Research and Development Program of China (2022YFC2703004 to J.W. and 2022YFA1304500 to X.F.). "Pioneer" and "Leading Goose" R&D program of Zhejiang (2023SDYXS0001 to X.F. and Y.J.) and TIPC Director’s Fund to Q.F. We thank G. Qin for the moral support and H. Zhang for the polymer characterization. We also thank L. Chen and K. Wang for their help with Hessian-SIM imaging. Supplementary Fig. 4a is created with BioRender.com.
Author information
Authors and Affiliations
Contributions
X.F. and J.W. designed the research. X.F., J.W., Q.F. and Y.J. supervised the project. H.Y., Z.Y., X.L. and W.L. performed the experiments. Z.Y. and Q.F. synthesized the particles. H.Y., Z.Y., P.L., X.L., Y.W., H.X. and Y.J. analysed the data. H.Y., Y.J., Q.F., J.W. and X.F. wrote the paper. H.Y., Q.F. and J.W. drew Figs. 1a and 5a. All the authors contributed to discussing and editing the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Photonics thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note, Supplementary Discussion, Figs. 1–27, and Tables 1 and 2.
Source data
Source Data Figs. 1–5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, H., Yan, Z., Liu, X. et al. Single-chain ultrasmall fluorescent polymer dots enable nanometre-resolution cellular imaging and single protein tracking. Nat. Photon. 19, 1336–1344 (2025). https://doi.org/10.1038/s41566-025-01767-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41566-025-01767-1
This article is cited by
-
Polymer dots for nanoscale live-cell imaging
Nature Photonics (2025)