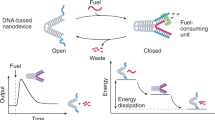
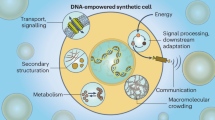
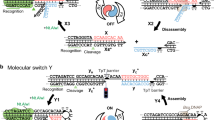
Abstract
DNA nanotechnology has rapidly evolved, leading to the development of dynamic nanoscale and microscale devices that mimic natural molecular machinery. This Review explores the latest advancements in DNA-based machines, motors and switches, emphasizing the need for clear definitions to distinguish between these often-interchanged terms. By analysing key performance metrics such as speed, force generation, efficiency and autonomy, we provide a framework for evaluating these devices against their biological counterparts, including motor proteins such as myosin and kinesin. We highlight innovative design strategies such as strand displacement, DNA origami and hybrid systems, which enhance the functionality of DNA-based constructs and bridge the gap between synthetic and natural systems. These advancements have promising applications in areas such as targeted drug delivery, biosensing and nanofabrication, although challenges in achieving the high performance and efficiency seen in biological systems remain. Through a synthesis of current research, this Review outlines the opportunities and challenges in the development of DNA-based nanoscale and microscale devices.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Schliwa, M. & Woehlke, G. Molecular motors. Nature 422, 759–765 (2003).
Vale, R. D. et al. Direct observation of single kinesin molecules moving along microtubules. Nature 380, 451–453 (1996).
Rayment, I. et al. Structure of the actin-myosin complex and its implications for muscle contraction. Science 261, 58–65 (1993).
Patel, S. S. & Donmez, I. Mechanisms of helicases. J. Biol. Chem. 281, 18265–18268 (2006).
Bhattacharyya, S., Yu, H., Mim, C. & Matouschek, A. Regulated protein turnover: snapshots of the proteasome in action. Nat. Rev. Mol. Cell Biol. 15, 122–133 (2014).
Richards, V. 2016 Nobel Prize in Chemistry: molecular machines. Nat. Chem. 8, 1090 (2016).
Linko, V. & Dietz, H. The enabled state of DNA nanotechnology. Curr. Opin. Biotechnol. 24, 555–561 (2013).
Dey, S. et al. DNA origami. Nat. Rev. Methods Prim. 1, 13 (2021).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Zhang, J. et al. Conditional deoxyribozyme-nanoparticle conjugates for miRNA-triggered gene regulation. ACS Appl. Mater. Interfaces 12, 37851–37861 (2020).
Narum, S. et al. An endosomal escape Trojan horse platform to improve cytosolic delivery of nucleic acids. ACS Nano 18, 6186–6201 (2024).
Li, S. et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264 (2018).
Velusamy, A., Sharma, R., Rashid, S. A., Ogasawara, H. & Salaita, K. DNA mechanocapsules for programmable piconewton responsive drug delivery. Nat. Commun. 15, 704 (2024).
McGhee, C. E., Loh, K. Y. & Lu, Y. DNAzyme sensors for detection of metal ions in the environment and imaging them in living cells. Curr. Opin. Biotechnol. 45, 191–201 (2017).
Liu, J. et al. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl Acad. Sci. USA 104, 2056–2061 (2007).
Thubagere, A. J. et al. A cargo-sorting DNA robot. Science https://doi.org/10.1126/science.aan6558 (2017).
Yehl, K. et al. High-speed DNA-based rolling motors powered by RNase H. Nat. Nanotechnol. 11, 184–190 (2016).
Blanchard, A. T. et al. Highly polyvalent DNA motors generate 100+ pN of force via autochemophoresis. Nano Lett. 19, 6977–6986 (2019).
Bazrafshan, A. et al. Tunable DNA origami motors translocate ballistically over μm distances at nm/s speeds. Angew. Chem. Int. Ed. 59, 9514–9521 (2020).
Bazrafshan, A. et al. DNA gold nanoparticle motors demonstrate processive motion with bursts of speed up to 50 nm per second. ACS Nano 15, 8427–8438 (2021).
Piranej, S., Bazrafshan, A. & Salaita, K. Chemical-to-mechanical molecular computation using DNA-based motors with onboard logic. Nat. Nanotechnol. 17, 514–523 (2022).
Zhang, L., Piranej, S., Namazi, A., Narum, S. & Salaita, K. “Turbo-charged” DNA motors with optimized sequence enable single-molecule nucleic acid sensing. Angew. Chem. Int. Ed. 63, e202316851 (2024).
Piranej, S. et al. Rolosense: mechanical detection of SARS-CoV-2 using a DNA-based motor. ACS Cent. Sci. 10, 1332–1347 (2024).
Piranej, S. et al. On-demand photoactivation of DNA-based motor motion. ACS Nano https://doi.org/10.1021/acsnano.4c13068 (2025).
Paxton, W. F. et al. Catalytic nanomotors: autonomous movement of striped nanorods. J. Am. Chem. Soc. 126, 13424–13431 (2004).
Dey, K. K. et al. Micromotors powered by enzyme catalysis. Nano Lett. 15, 8311–8315 (2015).
Wang, F., Liu, X. & Willner, I. DNA switches: from principles to applications. Angew. Chem. Int. Ed. 54, 1098–1129 (2015).
Fan, C., Plaxco, K. W. & Heeger, A. J. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl Acad. Sci. USA 100, 9134–9137 (2003).
Xiao, Y., Lai, R. Y. & Plaxco, K. W. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat. Protoc. 2, 2875–2880 (2007).
Tsong, T. T. Mechanisms of surface diffusion. Prog. Surf. Sci. 67, 235–248 (2001).
Li, J. et al. Exploring the speed limit of toehold exchange with a cartwheeling DNA acrobat. Nat. Nanotechnol. 13, 723–729 (2018).
Walker, D., Kubler, M., Morozov, K. I., Fischer, P. & Leshansky, A. M. Optimal length of low Reynolds number nanopropellers. Nano Lett. 15, 4412–4416 (2015).
Chen, S. et al. Nanomotors: 20 years anniversary and future roadmap. Preprint at ChemRxiv https://doi.org/10.26434/chemrxiv-2025-lczw9 (2025).
Peng, H., Li, X. F., Zhang, H. & Le, X. C. A microRNA-initiated DNAzyme motor operating in living cells. Nat. Commun. 8, 14378 (2017).
Tao, J., Zhang, H., Weinfeld, M. & Le, X. C. Development of a DNAzyme walker for the detection of APE1 in living cancer cells. Anal. Chem. 95, 14990–14997 (2023).
Korosec, C. S. et al. Motility of an autonomous protein-based artificial motor that operates via a burnt-bridge principle. Nat. Commun. 15, 1511 (2024).
Zhan, P., Jahnke, K., Liu, N. & Gopfrich, K. Functional DNA-based cytoskeletons for synthetic cells. Nat. Chem. 14, 958–963 (2022).
Alberts, B. et al. in Molecular Biology of the Cell Ch. 16, 4th edn (Garland Science, 2002).
Wijeratne, S. S., Fiorenza, S. A., Neary, A. E., Subramanian, R. & Betterton, M. D. Motor guidance by long-range communication on the microtubule highway. Proc. Natl Acad. Sci. USA 119, e2120193119 (2022).
Zhang, D. Y. & Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 (2011).
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Pal, N. Single-molecule FRET: a tool to characterize DNA nanostructures. Front. Mol. Biosci. 9, 835617 (2022).
Dai, Z., Xie, X., Gao, Z. & Li, Q. DNA-PAINT super-resolution imaging for characterization of nucleic acid nanostructures. ChemPlusChem 87, e202200127 (2022).
Du, Y., Pan, J. & Choi, J. H. A review on optical imaging of DNA nanostructures and dynamic processes. Methods Appl. Fluoresc. 7, 012002 (2019).
Endo, M. & Sugiyama, H. Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Acc. Chem. Res. 47, 1645–1653 (2014).
Kabiri, Y. et al. Distortion of DNA origami on graphene imaged with advanced TEM techniques. Small https://doi.org/10.1002/smll.201700876 (2017).
SantaLucia, J. Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA 95, 1460–1465 (1998).
Mao, C., Sun, W., Shen, Z. & Seeman, N. C. A nanomechanical device based on the B-Z transition of DNA. Nature 397, 144–146 (1999).
Cha, T.-G. et al. A synthetic DNA motor that transports nanoparticles along carbon nanotubes. Nat. Nanotechnol. 9, 39–43 (2014).
Centola, M. et al. A rhythmically pulsing leaf-spring DNA-origami nanoengine that drives a passive follower. Nat. Nanotechnol. 19, 226–236 (2024).
Qu, X. et al. An exonuclease III-powered, on-particle stochastic DNA walker. Angew. Chem. Int. Ed. 56, 1855–1858 (2017).
Zhou, L., Marras, A. E., Su, H.-J. & Castro, C. E. Direct design of an energy landscape with bistable DNA origami mechanisms. Nano Lett. 15, 1815–1821 (2015).
Gray, R. D., Li, J. & Chaires, J. B. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B 113, 2676–2683 (2009).
Harroun, S. G. et al. Programmable DNA switches and their applications. Nanoscale 10, 4607–4641 (2018).
Credi, A., Balzani, V., Langford, S. J. & Stoddart, J. F. Logic operations at the molecular level. An XOR gate based on a molecular machine. J. Am. Chem. Soc. 119, 2679–2681 (1997).
Hu, L., Lu, C.-H. & Willner, I. Switchable catalytic DNA catenanes. Nano Lett. 15, 2099–2103 (2015).
Lu, C.-H. et al. Switchable reconfiguration of a seven-ring interlocked DNA catenane nanostructure. Nano Lett. 15, 7133–7137 (2015).
Hu, Y., Ren, J., Lu, C.-H. & Willner, I. Programmed pH-driven reversible association and dissociation of interconnected circular DNA dimer nanostructures. Nano Lett. 16, 4590–4594 (2016).
Ackermann, D. et al. A double-stranded DNA rotaxane. Nat. Nanotechnol. 5, 436–442 (2010).
Willner, E. M. et al. Single-molecule observation of the photoregulated conformational dynamics of DNA origami nanoscissors. Angew. Chem. Int. Ed. 56, 15324–15328 (2017).
Kuzyk, A. et al. A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat. Commun. 7, 10591 (2016).
Parkinson, G. N., Lee, M. P. H. & Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417, 876–880 (2002).
Phan, A. T., Guéron, M. & Leroy, J.-L. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 299, 123–144 (2000).
Gehring, K., Leroy, J.-L. & Guéron, M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 363, 561–565 (1993).
Lannes, L., Halder, S., Krishnan, Y. & Schwalbe, H. Tuning the pH response of i-motif DNA oligonucleotides. ChemBioChem 16, 1647–1656 (2015).
Surana, S., Bhat, J. M., Koushika, S. P. & Krishnan, Y. An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nat. Commun. 2, 340 (2011).
Modi, S., Nizak, C., Surana, S., Halder, S. & Krishnan, Y. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 8, 459–467 (2013).
Leung, K., Chakraborty, K., Saminathan, A. & Krishnan, Y. A DNA nanomachine chemically resolves lysosomes in live cells. Nat. Nanotechnol. 14, 176–183 (2019).
Small, L. S. R. et al. The bar-hinge motor: a synthetic protein design exploiting conformational switching to achieve directional motility. N. J. Phys. 21, 013002 (2019).
Ala-Nissila, T., Ferrando, R. & Ying, S. C. Collective and single particle diffusion on surfaces. Adv. Phys. 51, 949–1078 (2002).
Kastantin, M. & Schwartz, D. K. Connecting rare DNA conformations and surface dynamics using single-molecule resonance energy transfer. ACS Nano 5, 9861–9869 (2011).
Schaich, M. A. & Van Houten, B. Searching for DNA damage: insights from single molecule analysis. Front. Mol. Biosci. https://doi.org/10.3389/fmolb.2021.772877 (2021).
Yin, P., Yan, H., Daniell, X. G., Turberfield, A. J. & Reif, J. H. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed. 43, 4906–4911 (2004).
Bath, J., Green, S. J. & Turberfield, A. J. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. 44, 4358–4361 (2005).
von Delius, M. & Leigh, D. A. Walking molecules. Chem. Soc. Rev. 40, 3656–3676 (2011).
Jung, C., Allen, P. B. & Ellington, A. D. A stochastic DNA walker that traverses a microparticle surface. Nat. Nanotechnol. 11, 157–163 (2016).
Yao, D. et al. Dynamic programming of a DNA walker controlled by protons. ACS Nano 14, 4007–4013 (2020).
Jung, C., Allen, P. B. & Ellington, A. D. A simple, cleated DNA walker that hangs on to surfaces. ACS Nano 11, 8047–8054 (2017).
Pei, R. et al. Behavior of polycatalytic assemblies in a substrate-displaying matrix. J. Am. Chem. Soc. 128, 12693–12699 (2006).
Omabegho, T., Sha, R. & Seeman, N. C. A bipedal DNA Brownian motor with coordinated legs. Science 324, 67–71 (2009).
Korosec, C. S. et al. Substrate stiffness tunes the dynamics of polyvalent rolling motors. Soft Matter 17, 1468–1479 (2021).
Harashima, T., Otomo, A. & Iino, R. Rational engineering of DNA-nanoparticle motor with high speed and processivity comparable to motor proteins. Nat. Commun. 16, 729 (2025).
Zhang, Y. & Hess, H. Chemically-powered swimming and diffusion in the microscopic world. Nat. Rev. Chem. 5, 500–510 (2021).
Ault, J. T. & Shin, S. Physicochemical hydrodynamics of particle diffusiophoresis driven by chemical gradients. Annu. Rev. Fluid Mech. 57, 227–255 (2025).
Blanchard, A. T., Piranej, S., Pan, V. & Salaita, K. Adhesive dynamics simulations of highly polyvalent DNA motors. J. Phys. Chem. B 126, 7495–7509 (2022).
Noji, H., Yasuda, R., Yoshida, M. & Kinosita, K. Jr. Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 (1997).
Berg, H. C. & Anderson, R. A. Bacteria swim by rotating their flagellar filaments. Nature 245, 380–382 (1973).
Ketterer, P., Willner, E. M. & Dietz, H. Nanoscale rotary apparatus formed from tight-fitting 3D DNA components. Sci. Adv. 2, e1501209 (2016).
van Delden, R. A. et al. Unidirectional molecular motor on a gold surface. Nature 437, 1337–1340 (2005).
Kopperger, E. et al. A self-assembled nanoscale robotic arm controlled by electric fields. Science 359, 296–301 (2018).
Pumm, A. K. et al. A DNA origami rotary ratchet motor. Nature 607, 492–498 (2022).
Shi, X. et al. Sustained unidirectional rotation of a self-organized DNA rotor on a nanopore. Nat. Phys. 18, 1105–1111 (2022).
Shi, X. et al. A DNA turbine powered by a transmembrane potential across a nanopore. Nat. Nanotechnol. 19, 338–344 (2024).
Peil, A. et al. DNA assembly of modular components into a rotary nanodevice. ACS Nano 16, 5284–5291 (2022).
Lin, M. et al. A magnetically powered nanomachine with a DNA clutch. Nat. Nanotechnol. 19, 646–651 (2024).
Breaker, R. R. & Joyce, G. F. A DNA enzyme that cleaves RNA. Chem. Biol. 1, 223–229 (1994).
Chandra, M., Sachdeva, A. & Silverman, S. K. DNA-catalyzed sequence-specific hydrolysis of DNA. Nat. Chem. Biol. 5, 718–720 (2009).
Santoro, S. W. & Joyce, G. F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA 94, 4262–4266 (1997).
Kim, H.-K. et al. Metal-dependent global folding and activity of the 8-17 DNAzyme studied by fluorescence resonance energy transfer. J. Am. Chem. Soc. 129, 6896–6902 (2007).
Bonaccio, M., Credali, A. & Peracchi, A. Kinetic and thermodynamic characterization of the RNA-cleaving 8-17 deoxyribozyme. Nucleic Acids Res. 32, 916–925 (2004).
Fang, S., Lee, H. J., Wark, A. W., Kim, H. M. & Corn, R. M. Determination of ribonuclease H surface enzyme kinetics by surface plasmon resonance imaging and surface plasmon fluorescence spectroscopy. Anal. Chem. 77, 6528–6534 (2005).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Srinivas, N. et al. On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic Acids Res. 41, 10641–10658 (2013).
Zhang, D. Y. & Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009).
Machinek, R. R., Ouldridge, T. E., Haley, N. E., Bath, J. & Turberfield, A. J. Programmable energy landscapes for kinetic control of DNA strand displacement. Nat. Commun. 5, 5324 (2014).
Zhang, D. Y., Turberfield, A. J., Yurke, B. & Winfree, E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 (2007).
Lund, K. et al. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Li, H. et al. A DNA molecular robot that autonomously walks on the cell membrane to drive cell motility. Angew. Chem. Int. Ed. 60, 26087–26095 (2021).
Zhang, Y. et al. Dynamic 3D DNA rolling walkers via directional movement on a lipid bilayer supported by Au@Fe3O4 nanoparticles for sensitive detection of miRNA-16. Anal. Chem. 94, 8346–8353 (2022).
Wen, Z.-B. et al. A dynamic 3D DNA nanostructure based on silicon-supported lipid bilayers: a highly efficient DNA nanomachine for rapid and sensitive sensing. Chem. Commun. 55, 13414–13417 (2019).
Pan, J. et al. Visible/near-infrared subdiffraction imaging reveals the stochastic nature of DNA walkers. Sci. Adv. 3, e1601600 (2017).
Pan, J. et al. Mimicking chemotactic cell migration with DNA programmable synthetic vesicles. Nano Lett. 19, 9138–9144 (2019).
Valero, J. & Škugor, M. Mechanisms, methods of tracking and applications of DNA walkers: a review. ChemPhysChem 21, 1971–1988 (2020).
Shin, J.-S. & Pierce, N. A. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 126, 10834–10835 (2004).
Sherman, W. B. & Seeman, N. C. A precisely controlled DNA biped walking device. Nano Lett. 4, 1203–1207 (2004).
Dufrêne, Y. F. et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 12, 295–307 (2017).
Wang, Z. L. & Lee, J. L. in Developments in Surface Contamination and Cleaning 2nd edn (eds Kohli, R. & Mittal, K. L.) 395–443 (William Andrew, 2008).
Wickham, S. F. J. et al. Direct observation of stepwise movement of a synthetic molecular transporter. Nat. Nanotechnol. 6, 166–169 (2011).
Wickham, S. F. J. et al. A DNA-based molecular motor that can navigate a network of tracks. Nat. Nanotechnol. 7, 169–173 (2012).
Gu, H., Chao, J., Xiao, S.-J. & Seeman, N. C. A proximity-based programmable DNA nanoscale assembly line. Nature 465, 202–205 (2010).
Umeda, K., McArthur, S. J. & Kodera, N. Spatiotemporal resolution in high-speed atomic force microscopy for studying biological macromolecules in action. Microscopy 72, 151–161 (2023).
Ando, T. et al. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl Acad. Sci. USA 98, 12468–12472 (2001).
Saxton, M. J. & Jacobson, K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399 (1997).
Michelotti, N., de Silva, C., Johnson-Buck, A. E., Manzo, A. J. & Walter, N. G. A bird’s eye view tracking slow nanometer-scale movements of single molecular nano-assemblies. Methods Enzymol. 475, 121–148 (2010).
Schnitzbauer, J., Strauss, M. T., Schlichthaerle, T., Schueder, F. & Jungmann, R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 12, 1198–1228 (2017).
Schueder, F. et al. An order of magnitude faster DNA-PAINT imaging by optimized sequence design and buffer conditions. Nat. Methods 16, 1101–1104 (2019).
Tomov, T. E. et al. DNA bipedal motor achieves a large number of steps due to operation using microfluidics-based interface. ACS Nano 11, 4002–4008 (2017).
Verbrugge, S., Lansky, Z. & Peterman, E. J. G. Kinesin’s step dissected with single-motor FRET. Proc. Natl Acad. Sci. USA 106, 17741–17746 (2009).
Yildiz, A. & Selvin, P. R. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc. Chem. Res. 38, 574–582 (2005).
Yildiz, A. et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 (2003).
Mo, D., Lakin, M. R. & Stefanovic, D. Logic circuits based on molecular spider systems. Biosystems 146, 10–25 (2016).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Chen, Z. et al. De novo design of protein logic gates. Science 368, 78–84 (2020).
Alberts, B., Wilson, J. H. & Hunt, T. Molecular Biology of the Cell 5th edn (Garland Science, 2008).
Wang, H. et al. A self-serviced-track 3D DNA walker for ultrasensitive detection of tumor exosomes by glycoprotein profiling. Angew. Chem. Int. Ed. 61, e202116932 (2022).
Miao, P., Chai, H. & Tang, Y. DNA hairpins and dumbbell-wheel transitions amplified walking nanomachine for ultrasensitive nucleic acid detection. ACS Nano 16, 4726–4733 (2022).
Luo, L. et al. Enhancing 3D DNA walker-induced CRISPR/Cas12a technology for highly sensitive detection of exomicroRNA associated with osteoporosis. ACS Sens. 9, 1438–1446 (2024).
Pan, M. C. et al. Wireframe orbit-accelerated bipedal DNA walker for electrochemiluminescence detection of methyltransferase activity. ACS Sens. 7, 2475–2482 (2022).
Zhang, Q. et al. A telomerase-assisted strategy for regeneration of DNA nanomachines in living cells. Angew. Chem. Int. Ed. 62, e202213884 (2023).
Guo, P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 5, 833–842 (2010).
Tran, M. P. et al. Genetic encoding and expression of RNA origami cytoskeletons in synthetic cells. Nat. Nanotechnol. https://doi.org/10.1038/s41565-025-01879-3 (2025).
Ibusuki, R. et al. Programmable molecular transport achieved by engineering protein motors to move on DNA nanotubes. Science 375, 1159–1164 (2022).
Chen, Y., Wang, M. & Mao, C. An autonomous DNA nanomotor powered by a DNA enzyme. Angew. Chem. Int. Ed. 43, 3554–3557 (2004).
Bath, J., Green, S. J., Allen, K. E. & Turberfield, A. J. Mechanism for a directional, processive, and reversible DNA motor. Small 5, 1513–1516 (2009).
Tomov, T. E. et al. Rational design of DNA motors: fuel optimization through single-molecule fluorescence. J. Am. Chem. Soc. 135, 11935–11941 (2013).
Zhou, C., Duan, X. & Liu, N. A plasmonic nanorod that walks on DNA origami. Nat. Commun. 6, 8102 (2015).
Zhan, P. et al. DNA-assembled nanoarchitectures with multiple components in regulated and coordinated motion. Sci. Adv. 5, eaax6023 (2019).
Liu, X. R. et al. A light-operated integrated DNA walker-origami system beyond bridge burning. Nanoscale Horiz. 8, 827–841 (2023).
Siti, W. et al. Autonomous DNA molecular motor tailor-designed to navigate DNA origami surface for fast complex motion and advanced nanorobotics. Sci. Adv. 9, eadi8444 (2023).
Anderson, T. et al. A light-powered single-stranded DNA molecular motor with colour-selective single-step control. Angew. Chem. Int. Ed. 63, e202405250 (2024).
Roke, D., Wezenberg, S. J. & Feringa, B. L. Molecular rotary motors: unidirectional motion around double bonds. Proc. Natl Acad. Sci. USA 115, 9423–9431 (2018).
Vale, R. D. The molecular motor toolbox for intracellular transport. Cell 112, 467–480 (2003).
Yin, P., Choi, H. M. T., Calvert, C. R. & Pierce, N. A. Programming biomolecular self-assembly pathways. Nature 451, 318–322 (2008).
Schnitzer, M. J., Visscher, K. & Block, S. M. Force production by single kinesin motors. Nat. Cell Biol. 2, 718–723 (2000).
Pierobon, P. et al. Velocity, processivity, and individual steps of single myosin V molecules in live cells. Biophys. J. 96, 4268–4275 (2009).
Ryan, J. M. & Nebenfuhr, A. Update on myosin motors: molecular mechanisms and physiological functions. Plant Physiol. 176, 119–127 (2018).
Okada, Y. & Hirokawa, N. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl Acad. Sci. USA 97, 640–645 (2000).
Reck-Peterson, S. L. et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell 126, 335–348 (2006).
Asbury, C. L., Fehr, A. N. & Block, S. M. Kinesin moves by an asymmetric hand-over-hand mechanism. Science 302, 2130–2134 (2003).
Toprak, E., Yildiz, A., Hoffman, M. T., Rosenfeld, S. S. & Selvin, P. R. Why kinesin is so processive. Proc. Natl Acad. Sci. USA 106, 12717–12722 (2009).
Blanchard, A. T. & Salaita, K. Multivalent molecular tension probes as anisotropic mechanosensors: concept and simulation. Phys. Biol. 18, 034001 (2021).
Hu, Y. et al. Quantifying T cell receptor mechanics at membrane junctions using DNA origami tension sensors. Nat. Nanotechnol. 19, 1674–1685 (2024).
Liu, Y., Galior, K., Ma, V. P. & Salaita, K. Molecular tension probes for imaging forces at the cell surface. Acc. Chem. Res. 50, 2915–2924 (2017).
Ma, R. et al. DNA probes that store mechanical information reveal transient piconewton forces applied by T cells. Proc. Natl Acad. Sci. USA 116, 16949–16954 (2019).
Ma, R. et al. Molecular mechanocytometry using tension-activated cell tagging. Nat. Methods 20, 1666–1671 (2023).
Zhang, Y., Ge, C., Zhu, C. & Salaita, K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat. Commun. 5, 5167 (2014).
Yasuda, R., Noji, H., Kinosita, K. Jr. & Yoshida, M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell 93, 1117–1124 (1998).
Toyabe, S., Watanabe-Nakayama, T., Okamoto, T., Kudo, S. & Muneyuki, E. Thermodynamic efficiency and mechanochemical coupling of F1-ATPase. Proc. Natl Acad. Sci. USA 108, 17951–17956 (2011).
Lau, A. W., Lacoste, D. & Mallick, K. Nonequilibrium fluctuations and mechanochemical couplings of a molecular motor. Phys. Rev. Lett. 99, 158102 (2007).
Vilfan, A. & Sarlah, A. Theoretical efficiency limits and speed-efficiency trade-off in myosin motors. PLoS Comput. Biol. 19, e1011310 (2023).
Ariga, T., Tomishige, M. & Mizuno, D. Nonequilibrium energetics of molecular motor kinesin. Phys. Rev. Lett. 121, 218101 (2018).
Wang, W., Chiang, T.-Y., Velegol, D. & Mallouk, T. E. Understanding the efficiency of autonomous nano- and microscale motors. J. Am. Chem. Soc. 135, 10557–10565 (2013).
Rempel, M. & Emberly, E. Optimizing efficiency and motility of a polyvalent molecular motor. Micromachines 13, 914 (2022).
Howard, J. Protein power strokes. Curr. Biol. 16, R517–R519 (2006).
Penocchio, E., Gu, G., Albaugh, A. & Gingrich, T. R. Power strokes in molecular motors: predictive, irrelevant, or somewhere in between? J. Am. Chem. Soc. 147, 1063–1073 (2025).
Barclay, C. J. in Muscle and Exercise Physiology (ed. Zoladz, J. A.) 111–127 (Academic, 2019).
Hongyun, W. & Oster, G. The Stokes efficiency for molecular motors and its applications. Europhys. Lett. 57, 134 (2002).
Li, C.-B. & Toyabe, S. Efficiencies of molecular motors: a comprehensible overview. Biophys. Rev. 12, 419–423 (2020).
Sarkar, A., LeVine, D. N., Kuzmina, N., Zhao, Y. & Wang, X. Cell migration driven by self-generated integrin ligand gradient on ligand-labile surfaces. Curr. Biol. 30, 4022–4032.e5 (2020).
Nakamura, A., Okazaki, K.-i., Furuta, T., Sakurai, M. & Iino, R. Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin. Nat. Commun. 9, 3814 (2018).
Hu, L., Vecchiarelli, A. G., Mizuuchi, K., Neuman, K. C. & Liu, J. Directed and persistent movement arises from mechanochemistry of the ParA/ParB system. Proc. Natl Acad. Sci. USA 112, E7055–E7064 (2015).
Hu, L., Vecchiarelli, A. G., Mizuuchi, K., Neuman, K. C. & Liu, J. Brownian ratchet mechanism for faithful segregation of low-copy-number plasmids. Biophys. J. 112, 1489–1502 (2017).
Vahey, M. D. & Fletcher, D. A. Influenza A virus surface proteins are organized to help penetrate host mucus. eLife 8, e43764 (2019).
Hamming, P. H., Overeem, N. J. & Huskens, J. Influenza as a molecular walker. Chem. Sci. 11, 27–36 (2020).
Feynman, R. P., Leighton, R. B. & Sands, M. L. The Feynman Lectures on Physics (Addison-Wesley, 1963).
Mai, J., Sokolov, I. M. & Blumen, A. Directed particle diffusion under “burnt bridges” conditions. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 64, 011102 (2001).
Acknowledgements
The authors acknowledge support from Merck Future Insight Prize (to K.S.) and Joachim Herz Stiftung Global Impact Award (to S.P.).
Author information
Authors and Affiliations
Contributions
S.P. and L.Z. contributed equally to this work. S.P. and L.Z. researched data for the article and prepared the figures. S.P. and A.B. discussed content and outlined topics. W.D. helped perform research for the article and worked on Table 1. S.P., L.Z. and K.S. discussed the content and wrote the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Chemistry thanks Hao Yan, Kerstin Göpfrich, Ryota Iino and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Burnt-bridge Brownian ratchet
-
(BBR). A molecular mechanism wherein motion is biased to a new foothold site by reducing the affinity to previously visited foothold sites through chemical or structural modifications.
- DNA diffusive walkers
-
DNA strands or supramolecular constructs that move randomly on a 2D surface as an effect of thermal fluctuations.
- DNA machines
-
Functional materials comprising DNA with the following features that mirror macroscopic machines: (1) consume chemical fuel to generate mechanical work, (2) processive and autonomous, (3) programmable and (4) responsive to inputs to perform functions such as cargo transport, sensing and drug release.
- DNA motors
-
Molecular machines made of DNA that operate far from equilibrium by consuming energy to generate active motion.
- DNA switches
-
Molecular constructs made of DNA that transition between two or more distinct states in response to an external input, such as light, pH change or force application.
- Efficiency
-
The figure of merit used to evaluate the performance of a molecular motor based on its capability to convert the energy input into mechanical work.
- Fuel
-
A molecule that is discarded or cleaved to allow a molecular motor to initiate a step by migrating to the successive attachment site.
- Hub
-
An inert body used by a molecular motor to coordinate the movement of multiple legs.
- Polyvalency
-
Also known as multivalency; the state or condition of a molecular motor that has multiple attachment contacts with its track at any given time.
- Processivity
-
The ability of a molecular motor to undertake multiple steps without dissociating from its track.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Piranej, S., Zhang, L., Bazrafshan, A. et al. Programming DNA machines to move. Nat Rev Chem (2026). https://doi.org/10.1038/s41570-025-00791-7
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41570-025-00791-7