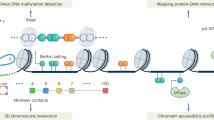
Abstract
DNA methylation is a critical epigenetic mechanism in numerous biological processes, including gene regulation, development, ageing and the onset of various diseases such as cancer. Studies of methylation are increasingly using single-molecule long-read sequencing technologies to simultaneously measure epigenetic states such as DNA methylation with genomic variation. These long-read data sets have spurred the continuous development of advanced computational methods to gain insights into the roles of methylation in regulating chromatin structure and gene regulation. In this Review, we discuss the computational methods for calling methylation signals, contrasting methylation between samples, analysing cell-type diversity and gaining additional genomic insights, and then further discuss the challenges and future perspectives of tool development for DNA methylation research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Moore, L. D., Le, T. & Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38 (2012).
Lucas, M. C. & Novoa, E. M. Long-read sequencing in the era of epigenomics and epitranscriptomics. Nat. Methods 20, 25–29 (2023).
Loyfer, N. et al. A DNA methylation atlas of normal human cell types. Nature 613, 355–364 (2023). This paper is the first to reveal the DNA methylation landscape of major normal human cell types.
Jones, M. J., Goodman, S. J. & Kobor, M. S. DNA methylation and healthy human aging. Aging Cell 14, 924 (2015).
Locke, W. J. et al. DNA methylation cancer biomarkers: translation to the clinic. Front. Genet. 10, 477856 (2019).
Ansar, M. et al. SMAD3 hypomethylation as a biomarker for early prediction of colorectal cancer. Int. J. Mol. Sci. 21, 7395 (2020).
Feinberg, A. P. & Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92 (1983).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome — biological and translational implications. Nat. Rev. Cancer 11, 726–734 (2011).
Ehrlich, M. DNA methylation in cancer: too much, but also too little. Oncogene 21, 5400–5413 (2002).
Berman, B. P. et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 44, 40–46 (2011).
Hansen, K. D. et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 43, 768–775 (2011).
Timp, W. & Feinberg, A. P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13, 497–510 (2013).
Si, W. et al. Nanopore sequencing identifies differentially methylated genes in the central nervous system in experimental autoimmune encephalomyelitis. J. Neuroimmunol. 381, 578134 (2023).
Ahmed, S. A. H., Ansari, S. A., Mensah-Brown, E. P. K. & Emerald, B. S. The role of DNA methylation in the pathogenesis of type 2 diabetes mellitus. Clin. Epigenetics 12, 104 (2020).
Johnson, A. A. et al. The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res. 15, 483 (2012).
Berdasco, M. & Esteller, M. Clinical epigenetics: seizing opportunities for translation. Nat. Rev. Genet. 20, 109–127 (2018).
Ahsan, M. U., Gouru, A., Chan, J., Zhou, W. & Wang, K. A signal processing and deep learning framework for methylation detection using Oxford Nanopore sequencing. Nat. Commun. 15, 1448 (2024).
Genner, R. et al. Assessing methylation detection for primary human tissue using Nanopore sequencing. Preprint at bioRxiv https://doi.org/10.1101/2024.02.29.581569 (2024).
Huang, Y.-W., Huang, T. H.-M. & Wang, L.-S. Profiling DNA methylomes from microarray to genome-scale sequencing. Technol. Cancer Res. Treat. 9, 139–147 (2010).
Clark, S. J., Statham, A., Stirzaker, C., Molloy, P. L. & Frommer, M. DNA methylation: bisulphite modification and analysis. Nat. Protoc. 1, 2353–2364 (2006).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868 (2005).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). This paper shows the importance of DMRs.
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Olova, N. et al. Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of sbiases affecting DNA methylation data. Genome Biol. 19, 33 (2018).
Sigurpalsdottir, B. D. et al. A comparison of methods for detecting DNA methylation from long-read sequencing of human genomes. Genome Biol. 25, 69 (2024).
Vaisvila, R. et al. Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res. 31, 1280–1289 (2021).
Liu, Y. et al. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol. 37, 424–429 (2019).
De Coster, W., Weissensteiner, M. H. & Sedlazeck, F. J. Towards population-scale long-read sequencing. Nat. Rev. Genet. 22, 572–587 (2021).
Dinh, H. Q. et al. Advanced methylome analysis after bisulfite deep sequencing: an example in Arabidopsis. PLoS ONE 7, e41528 (2012).
Johns Hopkins University, School of Medicine. Genetic Resources Core Facility. GRCF https://grcf.jhmi.edu/ (2022).
Noguera-Castells, A., García-Prieto, C. A., Álvarez-Errico, D. & Esteller, M. Validation of the new EPIC DNA methylation microarray (900K EPIC v2) for high-throughput profiling of the human DNA methylome. Epigenetics 18, 2185742 (2023).
Sedlazeck, F. J., Lee, H., Darby, C. A. & Schatz, M. C. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 19, 329–346 (2018).
Flusberg, B. A. et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465 (2010). This paper is the first to show that long-read sequencing is capable of detecting DNA methylation.
Timp, W., Comer, J. & Aksimentiev, A. DNA base-calling from a nanopore using a Viterbi algorithm. Biophys. J. 102, L37–L39 (2012).
Simpson, J. T. et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017). This paper presents the first widely adopted method (Nanopolish) capable of calling DNA methylation on ONT.
Karimzadeh, M., Ernst, C., Kundaje, A. & Hoffman, M. M. Umap and Bismap: quantifying genome and methylome mappability. Nucleic Acids Res. 46, e120 (2018).
Yuen, Z. W.-S. et al. Systematic benchmarking of tools for CpG methylation detection from nanopore sequencing. Nat. Commun. 12, 3438 (2021).
Liu, Y. et al. DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation. Genome Biol. 22, 295 (2021).
Mahmoud, M. et al. Structural variant calling: the long and the short of it. Genome Biol. 20, 246 (2019).
English, A. C. et al. Analysis and benchmarking of small and large genomic variants across tandem repeats. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02225-z (2024).
Martin, M. et al. WhatsHap: fast and accurate read-based phasing. Preprint at bioRxiv https://doi.org/10.1101/085050 (2016).
Logsdon, G. A., Vollger, M. R. & Eichler, E. E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 21, 597–614 (2020).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Suzuki, Y. et al. AgIn: measuring the landscape of CpG methylation of individual repetitive elements. Bioinformatics 32, 2911–2919 (2016).
Pacific Biosciences. Detecting DNA base modifications using single molecule, real-time sequencing White Paper, Base Modifications (PacBio, 2024).
Tse, O. Y. O. et al. Genome-wide detection of cytosine methylation by single molecule real-time sequencing. Proc. Natl Acad. Sci. USA 118, e2019768118 (2021).
Pacific Biosciences. Jasmine: predict 5mC in PacBio HiFi reads. GitHub https://github.com/PacificBiosciences/jasmine (2024).
Ni, P. et al. DNA 5-methylcytosine detection and methylation phasing using PacBio circular consensus sequencing. Nat. Commun. 14, 4054 (2023).
Vaswani, A. et al. Attention is all you need. In Proc. 31st International Conference on Neural Information Processing Systems (eds von Luxburg, U. et al.) 6000–6010 (Curran Associates, 2017).
Liu, Y., Liu, Z., Jiang, T., Zang, T. & Wang, Y. Comparison of the Nanopore and PacBio sequencing technologies for DNA 5-methylcytosine detection. In Proc. IEEE International Conference on Bioinformatics and Biomedicine 220–225 (IEEE, 2022).
Rand, A. C. et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 14, 411–413 (2017).
Oxford Nanopore Technology. megalodon. GitHub https://github.com/nanoporetech/megalodon (2024).
Liu, Q. et al. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 10, 2449 (2019).
Ni, P. et al. DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics 35, 4586–4595 (2019).
Bonet, J. et al. DeepMP: a deep learning tool to detect DNA base modifications on Nanopore sequencing data. Bioinformatics 38, 1235–1243 (2022).
Stanojević, D., Li, Z., Bakić, S., Foo, R. & Šikić, M. Rockfish: A transformer-based model for accurate 5-methylcytosine prediction from nanopore sequencing. Nat. Commun. 15, 5580 (2024).
Zhang, Y. et al. On the application of BERT models for nanopore methylation detection. In Proc. 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 320–327 (IEEE, 2021).
Yin, C. et al. NanoCon: contrastive learning-based deep hybrid network for nanopore methylation detection. Bioinformatics 40, btae046 (2024).
Oxford Nanopore Technology. dorado: Oxford Nanopore’s basecaller. GitHub https://github.com/nanoporetech/dorado (2024).
Gamaarachchi, H. et al. GPU accelerated adaptive banded event alignment for rapid comparative nanopore signal analysis. BMC Bioinform. 21, 343 (2020).
Tourancheau, A., Mead, E. A., Zhang, X.-S. & Fang, G. Discovering multiple types of DNA methylation from bacteria and microbiome using nanopore sequencing. Nat. Methods 18, 491–498 (2021).
He, Y. & Ecker, J. R. Non-CG methylation in the human genome. Annu. Rev. Genomics Hum. Genet. 16, 55 (2015).
Titcombe, P. et al. Human non-CpG methylation patterns display both tissue-specific and inter-individual differences suggestive of underlying function. Epigenetics 17, 653 (2022).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571 (2011).
Wu, X. & Zhang, Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 (2017).
Khare, T. et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon–intron boundary. Nat. Struct. Mol. Biol. 19, 1037–1043 (2012).
Pastor, W. A. et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473, 394–397 (2011).
Vasanthakumar, A. & Godley, L. A. 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet. 208, 167–177 (2015).
Pfeifer, G. P., Xiong, W., Hahn, M. A. & Jin, S.-G. The role of 5-hydroxymethylcytosine in human cancer. Cell Tissue Res. 356, 631 (2014).
Guo, X.-J. et al. Loss of 5-hydroxymethylcytosine induces chemotherapy resistance in hepatocellular carcinoma via the 5-hmC/PCAF/AKT axis. Cell Death Dis. 14, 79 (2023).
Sakamoto, Y. et al. Long-read whole-genome methylation patterning using enzymatic base conversion and nanopore sequencing. Nucleic Acids Res. 49, e81 (2021).
Sun, Z. et al. Nondestructive enzymatic deamination enables single-molecule long-read amplicon sequencing for the determination of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Genome Res. 31, 291–300 (2021).
Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 (2008).
Mastroeni, D. et al. Epigenetic changes in Alzheimer’s disease. Decrements in DNA methylation. Neurobiol. Aging 31, 2025–2037 (2010).
Mazzone, R. et al. The emerging role of epigenetics in human autoimmune disorders. Clin. Epigenetics 11, 34 (2019).
Akbari, V. et al. Megabase-scale methylation phasing using nanopore long reads and NanoMethPhase. Genome Biol. 22, 68 (2021).
Aryee, M. J. et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363 (2014).
Hansen, K. D., Langmead, B. & Irizarry, R. A. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 13, R83 (2012).
Feng, H. & Wu, H. Differential methylation analysis for bisulfite sequencing using DSS. Quant. Biol. 7, 327 (2019).
Oxford Nanopore Technologies. Modkit. ONT https://nanoporetech.github.io/modkit (2024).
Lee, I. et al. Simultaneous profiling of chromatin accessibility and methylation on human cell lines with nanopore sequencing. Nat. Methods 17, 1191–1199 (2020).
Vollger, M. R. et al. Segmental duplications and their variation in a complete human genome. Science 376, eabj6965 (2022).
Feng, H., Conneely, K. N. & Wu, H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 42, e69 (2014).
Piao, Y., Xu, W., Park, K. H., Ryu, K. H. & Xiang, R. Comprehensive evaluation of differential methylation analysis methods for bisulfite sequencing data. Int. J. Environ. Res. Public Health 18, 7975 (2021).
Akalin, A. et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012).
Mattei, G. et al. PoreMeth2: decoding the evolution of methylome alterations with Nanopore sequencing. Preprint at bioRxiv https://doi.org/10.1101/2024.10.03.616449 (2024).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
De Coster, W., Stovner, E. B. & Strazisar, M. methplotlib: analysis of modified nucleotides from nanopore sequencing. Bioinformatics 36, 3236–3238 (2020).
Gu, Z., Eils, R., Schlesner, M. & Ishaque, N. EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genomics 19, 234 (2018).
Li, Y., Ge, D. & Lu, C. The SMART App: an interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin 12, 71 (2019).
Cheetham, S. W., Kindlova, M. & Ewing, A. D. methylartist: tools for visualizing modified bases from nanopore sequence data. Bioinformatics 38, 3109 (2022).
Razaghi, R. et al. modbamtools: analysis of single-molecule epigenetic data for long-range profiling, heterogeneity, and clustering. Preprint at bioRxiv https://doi.org/10.1101/2022.07.07.499188 (2022).
Bodea, G. O. et al. LINE-1 retrotransposons contribute to mouse PV interneuron development. Nat. Neurosci. 27, 1274–1284 (2024).
Kolmogorov, M. et al. Scalable Nanopore sequencing of human genomes provides a comprehensive view of haplotype-resolved variation and methylation. Nat. Methods 20, 1483–1492 (2023).
Dolzhenko, E. et al. Characterization and visualization of tandem repeats at genome scale. Nat. Biotechnol. 42, 1606–1614 (2024).
Simmer, F. et al. Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics 7, 1355 (2012).
Sindi, S. et al. Promoter methylation-regulated differentially expressed genes in breast cancer. Breast Cancer 15, 435 (2023).
Gustafson, J. A. et al. High-coverage nanopore sequencing of samples from the 1000 Genomes Project to build a comprehensive catalog of human genetic variation. Genome Res. 34, 2061–2073 (2024).
Abante, J., Fang, Y., Feinberg, A. P. & Goutsias, J. Detection of haplotype-dependent allele-specific DNA methylation in WGBS data. Nat. Commun. 11, 5238 (2020).
Gigante, S. et al. Using long-read sequencing to detect imprinted DNA methylation. Nucleic Acids Res. 47, e46 (2019). This paper presents the first method to phase DNA methylation with SNP-based long-read phasing methods.
Nishiyama, A. & Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 37, 1012–1027 (2021).
Shoemaker, R., Deng, J., Wang, W. & Zhang, K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 20, 883 (2010).
Zhang, Y., Rohde, C., Reinhardt, R., Voelcker-Rehage, C. & Jeltsch, A. Non-imprinted allele-specific DNA methylation on human autosomes. Genome Biol. 10, R138 (2009).
Zink, F. et al. Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat. Genet. 50, 1542–1552 (2018).
Rosenski, J. et al. Atlas of imprinted and allele-specific DNA methylation in the human body. Preprint at bioRxiv https://doi.org/10.1101/2024.05.01.591988 (2024).
Fraser, H. B., Lam, L. L., Neumann, S. M. & Kobor, M. S. Population-specificity of human DNA methylation. Genome Biol. 13, R8 (2012).
Majidian, S. & Sedlazeck, F. J. PhaseME: automatic rapid assessment of phasing quality and phasing improvement. Gigascience 9, giaa078 (2020).
Hofmeister, R. J., Ribeiro, D. M., Rubinacci, S. & Delaneau, O. Accurate rare variant phasing of whole-genome and whole-exome sequencing data in the UK Biobank. Nat. Genet. 55, 1243–1249 (2023).
Kong, A. et al. Parental origin of sequence variants associated with complex diseases. Nature 462, 868–874 (2009).
Fu, Y. et al. MethPhaser: methylation-based long-read haplotype phasing of human genomes. Nat. Commun. 15, 5327 (2024). This work is the first study using long-read DNA methylation signals to enhance genome phasing.
Zhao, T., Hu, Y., Zang, T. & Wang, Y. Integrate GWAS, eQTL, and mQTL data to identify Alzheimer’s disease-related genes. Front. Genet. 10, 467372 (2019).
Ma, J. et al. Elucidating the genetic architecture of DNA methylation to identify promising molecular mechanisms of disease. Sci. Rep. 12, 19564 (2022).
Fan, Y. et al. IMAGE: high-powered detection of genetic effects on DNA methylation using integrated methylation QTL mapping and allele-specific analysis. Genome Biol. 20, 220 (2019).
Stefansson, O. A. et al. The correlation between CpG methylation and gene expression is driven by sequence variants. Nat. Genet. 56, 1624–1631 (2024). This important paper uses long reads to perform an mQTL study on an Icelandic population with the consideration of parent-of-origin DNA methylation.
Zhang, Y. et al. Global impact of somatic structural variation on the DNA methylome of human cancers. Genome Biol. 20, 209 (2019).
Katsman, E. et al. Detecting cell-of-origin and cancer-specific methylation features of cell-free DNA from Nanopore sequencing. Genome Biol. 23, 158 (2022).
Zhu, T. et al. A pan-tissue DNA methylation atlas enables in silico decomposition of human tissue methylomes at cell-type resolution. Nat. Methods 19, 296–306 (2022).
Houseman, E. A. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 13, 86 (2012).
Jaffe, A. E. & Irizarry, R. A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15, R31 (2014).
Schmidt, M., Maié, T., Dahl, E., Costa, I. G. & Wagner, W. Deconvolution of cellular subsets in human tissue based on targeted DNA methylation analysis at individual CpG sites. BMC Biol. 18, 178 (2020).
Yang, J. et al. DNA methylation-based epigenetic signatures predict somatic genomic alterations in gliomas. Nat. Commun. 13, 4410 (2022).
Zong, W. et al. scMethBank: a database for single-cell whole genome DNA methylation maps. Nucleic Acids Res. 50, D380–D386 (2021).
Capper, D. et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018). This paper describes the ability of using DNA methylation to classify central nervous system tumours.
Moss, J. et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018).
Oliva, M. et al. DNA methylation QTL mapping across diverse human tissues provides molecular links between genetic variation and complex traits. Nat. Genet. 55, 112–122 (2022).
Weinstein, J. N. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Nichols, R. V. et al. High-throughput robust single-cell DNA methylation profiling with sciMETv2. Nat. Commun. 13, 7627 (2022).
Luo, C. et al. Robust single-cell DNA methylome profiling with snmC-seq2. Nat. Commun. 9, 3824 (2018).
Franzen, J. et al. DNA methylation changes during long-term in vitro cell culture are caused by epigenetic drift. Commun. Biol. 4, 598 (2021).
Zhang, H., Cai, R., Dai, J. & Sun, W. EMeth: an EM algorithm for cell type decomposition based on DNA methylation data. Sci. Rep. 11, 5717 (2021).
Arneson, D., Yang, X. & Wang, K. MethylResolver — a method for deconvoluting bulk DNA methylation profiles into known and unknown cell contents. Commun. Biol. 3, 422 (2020).
Zhang, W. et al. ARIC: accurate and robust inference of cell type proportions from bulk gene expression or DNA methylation data. Brief. Bioinform. 23, bbab362 (2021).
Broadbent, J. nanomix. GitHub https://github.com/Jonbroad15/nanomix (2024).
Hannon, E. & Mill, J. Leveraging epigenetic signatures to determine the cell-type of origin from long read sequencing data. Preprint at bioRxiv https://doi.org/10.1101/2024.06.03.597114 (2024).
De Ridder, K., Che, H., Leroy, K. & Thienpont, B. Benchmarking of methods for DNA methylome deconvolution. Nat. Commun. 15, 4134 (2024).
Houseman, E. A. et al. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinform. 17, 259 (2016).
Chakravarthy, A. et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat. Commun. 9, 3220 (2018).
Lee, D., Lee, S. & Kim, S. PRISM: methylation pattern-based, reference-free inference of subclonal makeup. Bioinformatics 35, i520–i529 (2019).
Rahmani, E. et al. BayesCCE: a Bayesian framework for estimating cell-type composition from DNA methylation without the need for methylation reference. Genome Biol. 19, 141 (2018).
Titus, A. J., Gallimore, R. M., Salas, L. A. & Christensen, B. C. Cell-type deconvolution from DNA methylation: a review of recent applications. Hum. Mol. Genet. 26, R216 (2017).
Xie, H. et al. Genome-wide quantitative assessment of variation in DNA methylation patterns. Nucleic Acids Res. 39, 4099–4108 (2011).
Fang, Y. et al. DNA methylation entropy is associated with DNA sequence features and developmental epigenetic divergence. Nucleic Acids Res 51, 2046–2065 (2023).
Lee, D., Koo, B., Yang, J. & Kim, S. Metheor: ultrafast DNA methylation heterogeneity calculation from bisulfite read alignments. PLoS Comput. Biol. 19, e1010946 (2023).
Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26, 813–825 (2014).
Jenkinson, G., Pujadas, E., Goutsias, J. & Feinberg, A. P. Potential energy landscapes identify the information-theoretic nature of the epigenome. Nat. Genet. 49, 719–729 (2017).
Vaidya, H. et al. DNA methylation entropy as a measure of stem cell replication and aging. Genome Biol. 24, 27 (2023).
Lee, D. L. & Koo, B. K. DNA methylation heterogeneity profiles of 928 CCLE cell lines. figshare https://doi.org/10.6084/m9.figshare.21100717.v1 (2022).
Scherer, M. et al. Quantitative comparison of within-sample heterogeneity scores for DNA methylation data. Nucleic Acids Res. 48, e46 (2020).
Abante, J., Kambhampati, S., Feinberg, A. P. & Goutsias, J. Estimating DNA methylation potential energy landscapes from nanopore sequencing data. Sci. Rep. 11, 21619 (2021).
Kerr, L., Kafetzopoulos, I., Grima, R. & Sproul, D. Genome-wide single-molecule analysis of long-read DNA methylation reveals heterogeneous patterns at heterochromatin that reflect nucleosome organisation. PLoS Genet. 19, e1010958 (2023).
Villicaña, S. & Bell, J. T. Genetic impacts on DNA methylation: research findings and future perspectives. Genome Biol. 22, 127 (2021).
Bhat, A. et al. Role of transposable elements in genome stability: implications for health and disease. Int. J. Mol. Sci. 23, 7802 (2022).
Gershman, A. et al. Epigenetic patterns in a complete human genome. Science 376, eabj5089 (2022).
Hoyt, S. J. et al. From telomere to telomere: the transcriptional and epigenetic state of human repeat elements. Science 376, eabk3112 (2022). This important paper uses long reads to profile DNA methylation in human repeat elements.
Giesselmann, P. et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat. Biotechnol. 37, 1478–1481 (2019).
Ewing, A. D. et al. Nanopore sequencing enables comprehensive transposable element epigenomic profiling. Mol. Cell 80, 915–928.e5 (2020).
Ramirez, P. et al. Nanopore-based DNA long-read sequencing analysis of the aged human brain. Prerpint at bioRxiv https://doi.org/10.1101/2024.02.01.578450 (2024).
Miga, K. H. et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 585, 79–84 (2020).
Logsdon, G. A. et al. The variation and evolution of complete human centromeres. Nature 629, 136–145 (2024).
Schmidt, T. T. et al. High resolution long-read telomere sequencing reveals dynamic mechanisms in aging and cancer. Nat. Commun. 15, 5149 (2024).
Potapova, T. et al. Epigenetic control and inheritance of rDNA arrays. Preprint at bioRxiv https://doi.org/10.1101/2024.09.13.612795 (2024).
Rodriguez-Algarra, F. et al. Genetic variation at mouse and human ribosomal DNA influences associated epigenetic states. Genome Biol. 23, 54 (2022).
Stergachis, A. B., Debo, B. M., Haugen, E., Stirling Churchman, L. & Stamatoyannopoulos, J. A. Single-molecule regulatory architectures captured by chromatin fiber sequencing. Science 368, 1449–1454 (2020).
Jha, A. et al. DNA-m6A calling and integrated long-read epigenetic and genetic analysis with fibertools. Genome Res. 34, 1976–1986 (2024).
Hook, P. W. & Timp, W. Beyond assembly: the increasing flexibility of single-molecule sequencing technology. Nat. Rev. Genet. 24, 627–641 (2023). This important paper describes the application on single-molecule sequencing technologies towards epigenomics.
Mahmoud, M. et al. Utility of long-read sequencing for all of us. Nat. Commun. 15, 837 (2024).
Mei, X., Blanchard, J., Luellen, C., Conboy, M. J. & Conboy, I. M. Fail-tests of DNA methylation clocks, and development of a noise barometer for measuring epigenetic pressure of aging and disease. Aging 15, 8552–8575 (2023).
Olson, N. D. et al. Variant calling and benchmarking in an era of complete human genome sequences. Nat. Rev. Genet. 24, 464–483 (2023).
Liu, Z. et al. Author correction: towards accurate and reliable resolution of structural variants for clinical diagnosis. Genome Biol. 23, 198 (2022).
Khayat, M. M. et al. Hidden biases in germline structural variant detection. Genome Biol. 22, 347 (2021).
Liao, W.-W. et al. A draft human pangenome reference. Nature 617, 312–324 (2023).
Kovaka, S. et al. Uncalled4 improves nanopore DNA and RNA modification detection via fast and accurate signal alignment. Preprint at bioRxiv https://doi.org/10.1101/2024.03.05.583511 (2024).
Oxford Nanopore Technologies. Guppy protocol. ONT https://nanoporetech.com/document/Guppy-protocol#gns.searchValue=guppy (2018).
Kong, Y. et al. Critical assessment of DNA adenine methylation in eukaryotes using quantitative deconvolution. Science 375, 515–522 (2022).
Park, Y. & Wu, H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 32, 1446–1453 (2016).
Pacific Biosciences. MethBat: a battery of methylation tools for PacBio HiFi reads. GitHub https://github.com/PacificBiosciences/MethBat (2024).
De Coster, W. methplotlib. GitHub https://github.com/wdecoster/methplotlib (2024).
Lau, B. T. et al. Single-molecule methylation profiles of cell-free DNA in cancer with nanopore sequencing. Genome Med. 15, 33 (2023).
Vermeulen, C. et al. Ultra-fast deep-learned CNS tumour classification during surgery. Nature 622, 842–849 (2023).
Zhou, W. InfiniumMethylation BeadChips annotation. GitHub https://zwdzwd.github.io/InfiniumAnnotation (2024).
Beaulaurier, J. et al. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat. Commun. 6, 7438 (2015).
Ni, P. et al. Genome-wide detection of cytosine methylations in plant from Nanopore data using deep learning. Nat. Commun. 12, 5976 (2021).
Klughammer, J. et al. Comparative analysis of genome-scale, base-resolution DNA methylation profiles across 580 animal species. Nat. Commun. 14, 232 (2023).
Agustinho, D. P. et al. Unveiling microbial diversity: harnessing long-read sequencing technology. Nat. Methods 21, 954–966 (2024).
Ziller, M. J., Hansen, K. D., Meissner, A. & Aryee, M. J. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat. Methods 12, 230–232 (2015).
Faulk, C. Genome skimming with nanopore sequencing precisely determines global and transposon DNA methylation in vertebrates. Genome Res. 33, 948–956 (2023).
Pacific Biosciences. pb-CpG-tools: collection of tools for the analysis of CpG data. GitHub https://github.com/PacificBiosciences/pb-CpG-tools (2024).
Hansen, K. et al. bsseq. Bioconductor https://doi.org/10.18129/B9.BIOC.BSSEQ (2017).
Global Alliance for Genomics & Health. HTS format specifications. Samtools https://samtools.github.io/hts-specs/ (2024).
Acknowledgements
The authors thank L. Paulin, K. Hansen, A. Wengler, C. Saunders and P. Rescheneder for discussions, and E. Dolzhenko for help with Fig. 2. Y.F. and F.J.S. are supported by National Institutes of Health (NIH) (1UG3NS132105-01, UM1DA058229 and 1U01HG011758-01). W.T. is supported by HG009190 (National Human Genome Research Institute (NHGRI)).
Author information
Authors and Affiliations
Contributions
All authors researched the literature and contributed substantially to discussion of the content; Y.F. wrote the article and all authors reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
F.J.S. receives research support from Illumina, Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT). W.T. has two patents (8,748,091 and 8,394,584) licensed to ONT. Y.F. declares no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks Duncan Sproul, Quentin Gouil and Kai Wang for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
6mASCOPE: https://github.com/fanglab/6mASCOPE
bsseq: https://rdrr.io/github/hansenlab/bsseq/man/read.modkit.html
ccsmeth: https://github.com/PengNi/ccsmeth
ccsmethphase: https://github.com/PengNi/ccsmethphase
cfDNA: https://github.com/billytcl/nanopore_cfDNA
cfNano: https://github.com/methylgrammarlab/cfdna-ont
CNS tumour tissue-type atlas: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE109381
DeepMod2: https://github.com/WGLab/DeepMod2
DeepSignal2: https://github.com/bioinfomaticsCSU/deepsignal
Dorado: https://github.com/nanoporetech/dorado
DSS R Package: https://bioconductor.org/packages/release/bioc/vignettes/DSS/inst/doc/DSS.html
fibertools: https://github.com/fiberseq/fibertools-rs
GTEx: https://gtexportal.org/home/
Guppy: https://nanoporetech.com
Integrative Genomics Viewer (IGV): https://igv.org/
Jasmine: https://github.com/PacificBiosciences/jasmine
LongReadDNAmCTClassifier: https://github.com/ejh243/LongReadDNAmCTClassifier
MethBat: https://github.com/PacificBiosciences/MethBat
MethPhaser: https://github.com/treangenlab/methphaser
methplotlib: https://github.com/wdecoster/methplotlib
methylartist: https://github.com/adamewing/methylartist
modbamtools: https://github.com/rrazaghi/modbamtools
ModKit: https://nanoporetech.github.io/modkit/
NanoMethPhase: https://github.com/vahidAK/NanoMethPhase
Nanomix: https://github.com/Jonbroad15/nanomix
Nanopolish/f5c: https://github.com/jts/nanopolish
Normal human cell types atlas: https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186458
Pan-tissue DNA methylation atlas: https://github.com/aet21/EpiSCORE
pb-CpG-tools: https://github.com/PacificBiosciences/pb-CpG-tools
PoreMeth2: https://github.com/Lab-CoMBINE/PoreMeth2
Rockfish: https://github.com/lbcb-sci/rockfish
scMethBank: https://ngdc.cncb.ac.cn/methbank/scm/documentation
Sturgeon: https://github.com/marcpaga/sturgeon
TCGA Pan-Cancer atlas: https://portal.gdc.cancer.gov/
TLDR: https://github.com/adamewing/tldr
TRGT: https://github.com/PacificBiosciences/trgt
UNCALLED4: https://github.com/skovaka/uncalled4
Glossary
- Attention model
-
A machine learning method that determines the relative importance of each component in a sequence.
- Beta distribution
-
A continuous probability distribution defined from 0 to 1.
- Bivariate shifting level model
-
A statistical model used to analyse paired data with shifting levels.
- Genomic imprinting
-
A process of gene silencing through DNA methylation — the repressed allele is methylated whereas the active allele is unmethylated.
- Graph genomes
-
A representation of genomic variation using a graph structure.
- Haplotype phasing
-
The process of determining which variants are located on the same chromosome copy (that is, haplotype).
- Hypermethylation
-
The increase in DNA methylation levels at specific genomic regions, often associated with gene silencing and implicated in various diseases, including cancer.
- Hypomethylation
-
The decrease in DNA methylation levels, which can lead to genomic instability, overexpression of oncogenes or reactivation of transposable elements.
- k-mer
-
A substring of length k from a DNA, RNA or protein sequence.
- Long short-term memory model
-
A specialized type of recurrent neural network (RNN) capable of learning long-term dependencies by incorporating a memory cell structure, mitigating the vanishing gradient problem.
- Non-negative matrix factorization
-
A dimensionality reduction technique that decomposes a matrix into non-negative components.
- Phase block
-
A contiguous genomic region where haplotypes are resolved.
- Recurrent neural networks
-
(RNNs). A class of neural network designed to process sequential data by maintaining a memory of past inputs, commonly used in time-series analysis and text prediction.
- Transformer architectures
-
Deep learning models that leverage self-attention mechanisms to process sequences in parallel, achieving state-of-the-art performance in tasks such as language modelling and sequence alignment.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fu, Y., Timp, W. & Sedlazeck, F.J. Computational analysis of DNA methylation from long-read sequencing. Nat Rev Genet 26, 620–634 (2025). https://doi.org/10.1038/s41576-025-00822-5
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41576-025-00822-5
This article is cited by
-
Non-CG DNA methylation in animal genomes
Nature Genetics (2025)