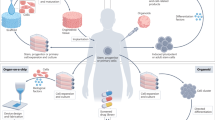
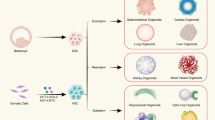
Abstract
Organoids are in vitro miniaturized and simplified model systems of organs that have gained enormous interest for modelling tissue development and disease, and for personalized medicine, drug screening and cell therapy. Despite considerable success in culturing physiologically relevant organoids, challenges remain to achieve real-life applications. In particular, the high variability of self-organizing growth and restricted experimental and analytical access hamper the translatability of organoid systems. In this Review, we argue that many limitations of traditional organoid culture can be addressed by engineering approaches at all levels of organoid systems. We investigate cell surface and genetic engineering approaches, and discuss stem cell niche engineering based on the design of matrices that allow spatiotemporal control of organoid growth and shape-guided morphogenesis. We examine how microfluidic approaches and lessons learnt from organs-on-a-chip enable the integration of mechano-physiological parameters and increase accessibility of organoids to improve functional readouts. Applying engineering principles to organoids increases reproducibility and provides experimental control, which will, ultimately, be required to enable clinical translation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Fatehullah, A., Tan, S. H. & Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–254 (2016).
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Rossi, G., Manfrin, A. & Lutolf, M. P. Progress and potential in organoid research. Nat. Rev. Genet. 19, 671–687 (2018).
Lancaster, M. A. & Knoblich, J. A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 (2014).
Lancaster, M. A. & Huch, M. Disease modelling in human organoids. Dis. Model. Mech. 12, dmm039347 (2019).
Kim, J., Koo, B. K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
McCauley, H. A. & Wells, J. M. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 144, 958–962 (2017).
Ootani, A. et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15, 701–706 (2009).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). This groundbreaking study describes the isolation of intestinal stem cells and showed that, in 3D culture, these stem cells can self-organize into epithelia with crypt-villus architecture featuring key cell types of the native tissue.
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). This study demonstrates the first generation of small intestinal and colon organoids from human adult tissue.
Jung, P. et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 17, 1225–1227 (2011).
Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–110 (2011). This work provides the first method for efficiently directing the differentiation of human pluripotent stem cells into patterned 3D intestinal tissue in vitro.
Workman, M. J. et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23, 49–59 (2017).
Tsai, Y. H. et al. In vitro patterning of pluripotent stem cell-derived intestine recapitulates in vivo human development. Development 144, 1045–1055 (2017).
Barker, N. et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Stange, D. E. et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155, 357–368 (2013).
Bartfeld, S. et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136.e6 (2015).
Bertaux-Skeirik, N. et al. CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation. PLoS Pathog. 11, e1004663 (2015).
McCracken, K. W. et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2014).
Noguchi, T. A. K. et al. Generation of stomach tissue from mouse embryonic stem cells. Nat. Cell Biol. 17, 984–993 (2015).
McCracken, K. W. et al. Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature 541, 182–187 (2017).
Taguchi, A. et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67 (2014).
Takasato, M. et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 16, 118–126 (2014).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Taguchi, A. & Nishinakamura, R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell 21, 730–746.e6 (2017).
Morizane, R. et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015).
Freedman, B. S. et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 8715 (2015).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Hu, H. et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606.e19 (2018).
Peng, W. C. et al. Inflammatory cytokine TNFα promotes the long-term expansion of primary hepatocytes in 3D culture. Cell 175, 1607–1619.e15 (2018).
Sampaziotis, F. et al. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat. Med. 23, 954–963 (2017).
Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013).
Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538 (2017).
Ang, L. T. et al. A roadmap for human liver differentiation from pluripotent stem cells. Cell Rep. 22, 2190–2205 (2018).
Takebe, T. et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 21, 2661–2670 (2017).
Sampaziotis, F. et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat. Biotechnol. 33, 845–852 (2015).
Ogawa, M. et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat. Biotechnol. 33, 853–861 (2015).
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013).
Greggio, C. et al. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development 140, 4452–4462 (2013).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Huang, L. et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21, 1364–1371 (2015).
Hohwieler, M. et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 66, 473–486 (2017).
Linnemann, J. R. et al. Quantification of regenerative potential in primary human mammary epithelial cells. Development 142, 3239–3251 (2015).
Jardé, T. et al. Wnt and Neuregulin1/ErbB signalling extends 3D culture of hormone responsive mammary organoids. Nat. Commun. 7, 13207 (2016).
Zhang, L. et al. Establishing estrogen-responsive mouse mammary organoids from single Lgr5+ cells. Cell. Signal. 29, 41–51 (2017).
Jamieson, P. R. et al. Derivation of a robust mouse mammary organoid system for studying tissue dynamics. Development 144, 1065–1071 (2017).
Xin, L., Lukacs, R. U., Lawson, D. A., Cheng, D. & Witte, O. N. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cell 25, 2760–2769 (2007).
Karthaus, W. R. et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175 (2014).
Höfner, T. et al. Defined conditions for the isolation and expansion of basal prostate progenitor cells of mouse and human origin. Stem Cell Rep. 4, 503–518 (2015).
Drost, J. et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 11, 347–358 (2016).
Tadokoro, T. et al. IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc. Natl Acad. Sci. USA 111, 3641–3649 (2014).
Jain, R. et al. Plasticity of Hopx+ type I alveolar cells to regenerate type II cells in the lung. Nat. Commun. 6, 6727 (2015).
Nikolic´, M. Z. et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. eLife 6, e26575 (2017).
Sachs, N. et al. Long- term expanding human airway organoids for disease modeling. EMBO J. 38, e100300 (2019).
Wong, A. P. et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 30, 876–882 (2012).
Dye, B. R. et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 4, e05098 (2015).
Dye, B. R. et al. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. eLife 5, e19732 (2016).
Chen, Y.-W. et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19, 542–549 (2017).
Antonica, F. et al. Generation of functional thyroid from embryonic stem cells. Nature 491, 66–71 (2012).
Kurmann, A. A. et al. Regeneration of thyroid function by transplantation of differentiated pluripotent stem cells. Cell Stem Cell 17, 527–542 (2015).
Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–58 (2011). This groundbreaking study describes the generation of embryonic stem-cell-derived retinal epithelia that self-pattern into neural retina and pigment epithelium domains.
Nakano, T. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012).
Phillips, M. J. et al. Blood-derived human iPS cells generate optic vesicle–like structures with the capacity to form retinal laminae and develop synapses. Investig. Opthalmol. Vis. Sci. 53, 2007–2019 (2012).
Reichman, S. et al. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc. Natl Acad. Sci. USA 111, 8518–8523 (2014).
Zhong, X. et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 5, 4047 (2014).
Völkner, M. et al. Retinal organoids from pluripotent stem cells efficiently recapitulate retinogenesis. Stem Cell Rep. 6, 525–538 (2016).
Capowski, E. E. et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 146, dev171686 (2019).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). This work constitutes the first report of the generation of cerebral organoids from human pluripotent stem cells.
Camp, J. G. et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl Acad. Sci. USA 112, 15672–15677 (2015).
Bagley, J. A., Reumann, D., Bian, S., Lévi-Strauss, J. & Knoblich, J. A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 14, 743–751 (2017).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Xiang, Y. et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell 21, 383–398.e7 (2017).
Kadoshima, T. et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl Acad. Sci. USA 110, 20284–20289 (2013).
Mariani, J. et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390 (2015).
Sakaguchi, H. et al. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 6, 8896 (2015).
Pas˛ca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Qian, X. et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016).
Jo, J. et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 19, 248–257 (2016).
Muguruma, K., Nishiyama, A., Kawakami, H., Hashimoto, K. & Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 10, 537–550 (2015).
Qian, X. et al. Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 13, 565–580 (2018).
Watson, L. M., Wong, M. M. K., Vowles, J., Cowley, S. A. & Becker, E. B. E. A simplified method for generating purkinje cells from human-induced pluripotent stem cells. Cerebellum 17, 419–427 (2018).
Eiraku, M. et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532 (2008).
Zachos, N. C. et al. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J. Biol. Chem. 291, 3759–3766 (2016).
Di Lullo, E. & Kriegstein, A. R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 18, 573–584 (2017).
Qian, X., Song, H. & Ming, G. L. Brain organoids: advances, applications and challenges. Development 146, dev166074 (2019).
Smits, L. M. et al. Modeling Parkinson’s disease in midbrain-like organoids. NPJ Parkinsons Dis. 5, 5 (2019).
Monzel, A. S. et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Rep. 8, 1144–1154 (2017).
Assawachananont, J. et al. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Rep. 2, 662–674 (2014).
Rashidi, H. et al. 3D human liver tissue from pluripotent stem cells displays stable phenotype in vitro and supports compromised liver function in vivo. Arch. Toxicol. 92, 3117–3129 (2018).
Blackford, S. J. I. et al. Validation of current good manufacturing practice compliant human pluripotent stem cell-derived hepatocytes for cell-based therapy. Stem Cell Transl. Med. 8, 124–137 (2019).
Lu, D. & Kassab, G. S. Role of shear stress and stretch in vascular mechanobiology. J. R. Soc. Interface 8, 1379–1385 (2011).
Abraham, G. et al. Growth and differentiation of primary and passaged equine bronchial epithelial cells under conventional and air-liquid-interface culture conditions. BMC Vet. Res. 7, 26 (2011).
Park, J. S. et al. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials 32, 3921–3930 (2011).
Park, S. E., Georgescu, A. & Huh, D. Organoids-on-a-chip. Science 364, 960–965 (2019).
Velasco, S. et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527 (2019).
Kim, G. A., Ginga, N. J. & Takayama, S. Integration of sensors in gastrointestinal organoid culture for biological analysis. Cell. Mol. Gastroenterol. Hepatol. 6, 123–131.e1 (2018).
Collins, S. D. et al. in Hepatocellular Carcinoma Ch. 3 (ed. Tirnitz-Parker, J. E. E.) 47–67 (Codon Publications, 2019).
Brassard, J. A. & Lutolf, M. P. Engineering stem cell self-organization to build better organoids. Cell Stem Cell 24, 860–876 (2019).
Fujimichi, Y., Otsuka, K., Tomita, M. & Iwasaki, T. An efficient intestinal organoid system of direct sorting to evaluate stem cell competition in vitro. Sci. Rep. 9, 20297 (2019).
Amaral, A. J. R. & Pasparakis, G. Cell membrane engineering with synthetic materials: Applications in cell spheroids, cellular glues and microtissue formation. Acta Biomater. 90, 21–36 (2019).
Rogozhnikov, D., O’Brien, P. J., Elahipanah, S. & Yousaf, M. N. Scaffold free bio-orthogonal assembly of 3-dimensional cardiac tissue via cell surface engineering. Sci. Rep. 6, 39806 (2016).
O’Brien, P. J., Luo, W., Rogozhnikov, D., Chen, J. & Yousaf, M. N. Spheroid and tissue assembly via click chemistry in microfluidic flow. Bioconjug. Chem. 26, 1939–1949 (2015).
Luo, W., Pulsipher, A., Dutta, D., Lamb, B. M. & Yousaf, M. N. Remote control of tissue interactions via engineered photo-switchable cell surfaces. Sci. Rep. 4, 6313 (2014).
Akbari, E. et al. Engineering cell surface function with DNA origami. Adv. Mater. 29, 1703632 (2017).
Fayol, D. et al. Use of magnetic forces to promote stem cell aggregation during differentiation, and cartilage tissue modeling. Adv. Mater. 25, 2611–2616 (2013).
Xia, P. F., Ling, H., Foo, J. L. & Chang, M. W. Synthetic genetic circuits for programmable biological functionalities. Biotechnol. Adv. 37, 107393 (2019).
Bashor, C. J. & Collins, J. J. Understanding biological regulation through synthetic biology. Annu. Rev. Biophys. 47, 399–423 (2018).
Zhang, F. Development of CRISPR-Cas systems for genome editing and beyond. Q. Rev. Biophys. 52, e6 (2019).
Velazquez, J. J. et al. Gene regulatory network analysis and engineering directs development and vascularization of multilineage human liver organoids. Cell Syst. 12, 41–55 (2020).
Roper, J. & Yilmaz, Ö. H. Breakthrough moments: genome editing and organoids. Cell Stem Cell 24, 841–842 (2019).
Schwank, G. et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658 (2013).
Deng, W. L. et al. Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Rep. 10, 1267–1281 (2018).
Matano, M. et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 (2015).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015).
Li, X. et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 20, 769–777 (2014).
Sekine, R., Shibata, T. & Ebisuya, M. Synthetic mammalian pattern formation driven by differential diffusivity of Nodal and Lefty. Nat. Commun. 9, 5456 (2018).
Guye, P. et al. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 7, 10243 (2016).
Kunche, S., Yan, H., Calof, A. L., Lowengrub, J. S. & Lander, A. D. Feedback, lineages and self-organizing morphogenesis. PLoS Comput. Biol. 12, e1004814 (2016).
Morsut, L. et al. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell 164, 780–791 (2016).
Toda, S., Blauch, L. R., Tang, S. K. Y., Morsut, L. & Lim, W. A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 361, 156–162 (2018). This study reports the generation of self-organizing structures employing a purely synthetic biology approach.
Hartmann, J., Krueger, D. & De Renzis, S. Using optogenetics to tackle systems-level questions of multicellular morphogenesis. Curr. Opin. Cell Biol. 66, 19–27 (2020).
Bateman, J. F., Boot-Handford, R. P. & Lamandé, S. R. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat. Rev. Genet. 10, 173–183 (2009).
Cox, T. R. & Erler, J. T. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Model. Mech. 4, 165–178 (2011).
Kleinman, H. K. & Martin, G. R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 15, 378–386 (2005).
Aisenbrey, E. A. & Murphy, W. L. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 5, 539–551 (2020).
Kratochvil, M. J. et al. Engineered materials for organoid systems. Nat. Rev. Mater. 4, 606–622 (2019).
Broguiere, N. et al. Growth of epithelial organoids in a defined hydrogel. Adv. Mater. 30, 1801621 (2018).
Jabaji, Z. et al. Type I collagen as an extracellular matrix for the in vitro growth of human small intestinal epithelium. PLoS ONE 9, e107814 (2014).
Lindborg, B. A. et al. Rapid induction of cerebral organoids from human induced pluripotent stem cells using a chemically defined hydrogel and defined cell culture medium. Stem Cell Transl. Med. 5, 970–979 (2016).
Gjorevski, N. et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016).
Cruz-Acuña, R. et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 19, 1326–1335 (2017).
Ranga, A. et al. 3D niche microarrays for systems-level analyses of cell fate. Nat. Commun. 5, 4324 (2014).
Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Chaudhuri, O., Cooper-White, J., Janmey, P. A., Mooney, D. J. & Shenoy, V. B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 584, 535–546 (2020).
Chaudhuri, O. et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 15, 326–334 (2016).
Nelson, C. M., VanDuijn, M. M., Inman, J. L., Fletcher, D. A. & Bissell, M. J. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science 314, 298–300 (2006).
Wang, Y. et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 128, 44–55 (2017).
Nikolaev, M. et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585, 574–578 (2020). This work reports the controlled generation of long-lived intestinal organoids using scaffolds with predefined geometry.
Lancaster, M. A. et al. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659–666 (2017).
Karzbrun, E., Kshirsagar, A., Cohen, S. R., Hanna, J. H. & Reiner, O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 14, 515–522 (2018).
Kloxin, A. M., Kasko, A. M., Salinas, C. N. & Anseth, K. S. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 324, 59–63 (2009).
Mosiewicz, K. A. et al. In situ cell manipulation through enzymatic hydrogel photopatterning. Nat. Mater. 12, 1072–1078 (2013).
Ruskowitz, E. R. & Deforest, C. A. Photoresponsive biomaterials for targeted drug delivery and 4D cell culture. Nat. Rev. Mater. 3, 17087 (2018).
Attayek, P. J. et al. In vitro polarization of colonoids to create an intestinal stem cell compartment. PLoS ONE 11, e153795 (2016).
Tabata, Y. & Lutolf, M. P. Multiscale microenvironmental perturbation of pluripotent stem cell fate and self-organization. Sci. Rep. 7, 44711 (2017).
Demers, C. J. et al. Development-on-chip: In vitro neural tube patterning with a microfluidic device. Development 143, 1884–1892 (2016).
Manfrin, A. et al. Engineered signaling centers for the spatially controlled patterning of human pluripotent stem cells. Nat. Methods 16, 640–648 (2019).
Cederquist, G. Y. et al. Specification of positional identity in forebrain organoids. Nat. Biotechnol. 37, 436–444 (2019).
Wylie, R. G. et al. Spatially controlled simultaneous patterning of multiple growth factors in three-dimensional hydrogels. Nat. Mater. 10, 799–806 (2011).
Battista, S. et al. The effect of matrix composition of 3D constructs on embryonic stem cell differentiation. Biomaterials 26, 6194–6207 (2005).
Teller, I. C. et al. Laminins in the developing and adult human small intestine: Relation with the functional absorptive unit. Dev. Dyn. 236, 1980–1990 (2007).
Groulx, J. F. et al. Collagen VI is a basement membrane component that regulates epithelial cell–fibronectin interactions. Matrix Biol. 30, 195–206 (2011).
Benoit, Y. D., Groulx, J.-F., Gagné, D. & Beaulieu, J.-F. RGD-dependent epithelial cell-matrix interactions in the human intestinal Crypt. J. Signal Transduct. 2012, 248759 (2012).
Broguiere, N. et al. Morphogenesis guided by 3D patterning of growth factors in biological matrices. Adv. Mater. 32, 1908299 (2020).
Torgersen, J. et al. Hydrogels for two-photon polymerization: A toolbox for mimicking the extracellular matrix. Adv. Funct. Mater. 23, 4542–4554 (2013).
Pradhan, S., Keller, K. A., Sperduto, J. L. & Slater, J. H. Fundamentals of laser-based hydrogel degradation and applications in cell and tissue engineering. Adv. Healthc. Mater. 6, 1700681 (2017).
Mammoto, T. & Ingber, D. E. Mechanical control of tissue and organ development. Development 137, 1407–1420 (2010).
Vianello, S. & Lutolf, M. P. Understanding the mechanobiology of early mammalian development through bioengineered models. Dev. Cell 48, 751–763 (2019).
Heo, I. et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823 (2018).
Leslie, J. L. et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 83, 138–145 (2015).
Schumacher, M. A. et al. The use of murine-derived fundic organoids in studies of gastric physiology. J. Physiol. 593, 1809–1827 (2015).
Co, J. Y. et al. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 26, 2509–2520.e4 (2019).
Huh, D. et al. Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668 (2010). This groundbreaking work describes an organ-on-a-chip approach to emulate lung functions in vitro.
Kim, H. J., Huh, D., Hamilton, G. & Ingber, D. E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174 (2012).
Musah, S. et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat. Biomed. Eng. 1, 0069 (2017).
Akhtar, A. A., Sances, S., Barrett, R. & Breunig, J. J. Organoid and organ-on-a-chip systems: new paradigms for modeling neurological and gastrointestinal disease. Curr. Stem Cell Rep. 3, 98–111 (2017).
Gayer, C. P. & Basson, M. D. The effects of mechanical forces on intestinal physiology and pathology. Cell. Signal. 21, 1237–1244 (2009).
Lee, K. K. et al. Human stomach-on-a-chip with luminal flow and peristaltic-like motility. Lab Chip 18, 3079–3085 (2018).
Homan, K. A. et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262 (2019). This study exemplifies the application of fluid flow to kidney organoids to enhance their level of vascularization and maturation.
Kasendra, M. et al. Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci. Rep. 8, 2871 (2018).
Imura, Y., Sato, K. & Yoshimura, E. Micro total bioassay system for ingested substances: Assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal. Chem. 82, 9983–9988 (2010).
Miller, P. G. & Shuler, M. L. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol. Bioeng. 113, 2213–2227 (2016).
Jin, Y. et al. Vascularized liver organoids generated using induced hepatic tissue and dynamic liver-specific microenvironment as a drug testing platform. Adv. Funct. Mater. 28, 1801954 (2018).
Skardal, A. et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 7, 8837 (2017).
Garcez, P. P. et al. Zika virus: Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816–818 (2016).
Dang, J. et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell 19, 258–265 (2016).
Gabriel, E. et al. Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell 20, 397–406.e5 (2017).
Huang, J. Y. et al. Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell Host Microbe 18, 147–156 (2015).
Holokai, L. et al. Increased programmed death-ligand 1 is an early epithelial cell response to Helicobacter pylori infection. PLoS Pathog. 15, e1007468 (2019).
Burkitt, M. D., Duckworth, C. A., Williams, J. M. & Pritchard, D. M. Helicobacter pylori-induced gastric pathology: Insights from in vivo and ex vivo models. Dis. Model. Mech. 10, 89–104 (2017).
Mejías-Luque, R. & Gerhard, M. Immune evasion strategies and persistence of Helicobacter pylori. Curr. Top. Microbiol. Immunol. 400, 53–71 (2017).
Ye, W., Luo, C., Li, C., Huang, J. & Liu, F. Organoids to study immune functions, immunological diseases and immunotherapy. Cancer Lett. 477, 31–40 (2020).
Roh, T. T., Chen, Y., Paul, H. T., Guo, C. & Kaplan, D. L. 3D bioengineered tissue model of the large intestine to study inflammatory bowel disease. Biomaterials 225, 119517 (2019).
Czerniecki, S. M. et al. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 22, 929–940.e4 (2018).
Brandenberg, N. et al. High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat. Biomed. Eng. 4, 863–874 (2020).
Jin, B. J. et al. Microfluidics platform for measurement of volume changes in immobilized intestinal enteroids. Biomicrofluidics 8, 024106 (2014).
Wang, Y. et al. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip 18, 3606–3616 (2018).
Artegiani, B. et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat. Cell Biol. 22, 321–331 (2020).
Sun, D., Evans, L. D. & Rawlins, E. L. Organoid Easytag: an efficient workflow for gene targeting in human organoids. Preprint at bioRxiv 10.1101/2020.05.04.076067 (2020).
Okkelman, I. A., Foley, T., Papkovsky, D. B. & Dmitriev, R. I. Live cell imaging of mouse intestinal organoids reveals heterogeneity in their oxygenation. Biomaterials 146, 86–96 (2017).
Muta, Y. et al. Composite regulation of ERK activity dynamics underlying tumour-specific traits in the intestine. Nat. Commun. 9, 2174 (2018).
Lukonin, I. et al. Phenotypic landscape of intestinal organoid regeneration. Nature 586, 275–280 (2020).
Adam Kratz, S. R., Höll, G., Schuller, P., Ertl, P. & Rothbauer, M. Latest trends in biosensing for microphysiological organs-on-a-chip and body-on-a-chip systems. Biosensors 9, 110 (2019).
Zhang, Y. S. et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl Acad. Sci. USA 114, E2293–E2302 (2017). This work reports a multi-organ microfluidics platform with automated and continuous monitoring of cultured organoids.
Bavli, D. et al. Real-time monitoring of metabolic function in liver-onchip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl Acad. Sci. USA 113, E2231–E2240 (2016).
Misun, P. M., Rothe, J., Schmid, Y. R. F., Hierlemann, A. & Frey, O. Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsyst. Nanoeng. 2, 16022 (2016).
Weltin, A. et al. Accessing 3D microtissue metabolism: Lactate and oxygen monitoring in hepatocyte spheroids. Biosens. Bioelectron. 87, 941–948 (2017).
Taurino, I. et al. Platinum nanopetal-based potassium sensors for acute cell death monitoring. RSC Adv. 6, 40517–40526 (2016).
Caluori, G. et al. Non-invasive electromechanical cell-based biosensors for improved investigation of 3D cardiac models. Biosens. Bioelectron. 124–125, 129–135 (2019).
Shin, S. R. et al. Label-free and regenerative electrochemical microfluidic biosensors for continual monitoring of cell secretomes. Adv. Sci. 4, 1600522 (2017).
Liu, H. et al. A droplet microfluidic system to fabricate hybrid capsules enabling stem cell organoid engineering. Adv. Sci. 7, 1903739 (2020).
Wang, Y. et al. One-step synthesis of composite hydrogel capsules to support liver organoid generation from hiPSCs. Biomater. Sci. 8, 5476–5488 (2020).
Laperrousaz, B. et al. Direct transfection of clonal organoids in Matrigel microbeads: A promising approach toward organoid-based genetic screens. Nucleic Acids Res. 46, e70 (2018).
Dolega, M. E., Abeille, F., Picollet-D’hahan, N. & Gidrol, X. Controlled 3D culture in Matrigel microbeads to analyze clonal acinar development. Biomaterials 52, 347–357 (2015).
Sachs, N. et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386.e10 (2018).
Reid, J. A., Mollica, P. M., Bruno, R. D. & Sachs, P. C. Consistent and reproducible cultures of large-scale 3D mammary epithelial structures using an accessible bioprinting platform. Breast Cancer Res. 20, 122 (2018).
Daly, A. C., Davidson, M. D. & Burdick, J. A. 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun. 12, 753 (2021).
Ayan, B. et al. Aspiration-assisted bioprinting for precise positioning of biologics. Sci. Adv. 6, eaaw5111 (2020).
Kim, T. Y. et al. Directed fusion of cardiac spheroids into larger heterocellular microtissues enables investigation of cardiac action potential propagation via cardiac fibroblasts. PLoS ONE 13, e0196714 (2018).
Brassard, J. A., Nikolaev, M., Hübscher, T., Hofer, M. & Lutolf, M. P. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 20, 22–29 (2021).
Dekkers, J. F. et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 19, 939–945 (2013).
Cora, V. et al. A cleared view on retinal organoids. Cells 8, 391 (2019).
Li, Y. et al. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell 20, 385–396.e3 (2017).
Serra, D. et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72 (2019).
Dekkers, J. F. et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 14, 1756–1771 (2019).
Choi, S. H. et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278 (2014).
Qin, X. et al. Cell-type-specific signaling networks in heterocellular organoids. Nat. Methods 17, 335–342 (2020).
Kanton, S. et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019).
Khrameeva, E. et al. Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. Genome Res. 30, 776–789 (2020).
Rheinwald, J. G. & Green, H. Serial cultivation of strains of human epidermal keratinocytes in defined clonal and serum-free culture. J. Invest. Dermatol. 6, 331–344 (1975).
Li, M. L. et al. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc. Natl Acad. Sci. USA 84, 136–140 (1987).
Clevers, H. & Watt, F. M. Defining adult stem cells by function, not by phenotype. Annu. Rev. Biochem. 87, 1015–1027 (2018).
Takebe, T., Zhang, B. & Radisic, M. Synergistic engineering: organoids meet organs-on-a-chip. Cell Stem Cell 21, 297–300 (2017).
Artegiani, B. et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell 24, 927–943.e6 (2019).
Seino, T. et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22, 454–467.e6 (2018).
Dekkers, J. F. et al. Modeling breast cancer using CRISPR-Cas9-mediated engineering of human breast organoids. J. Natl Cancer Inst. 112, 540–544 (2020).
Zhang, W. et al. Cerebral organoid and mouse models reveal a RAB39b–PI3K–mTOR pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Genes Dev. 34, 580–597 (2020).
Park, J. et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 21, 941–951 (2018).
Kim, H. et al. Modeling G2019S-LRRK2 sporadic Parkinson’s disease in 3D midbrain organoids. Stem Cell Rep. 12, 518–531 (2019).
Suzuki, K., Murano, T., Shimizu, H. & Ito, G. Single cell analysis of Crohn’ s disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J. Gastroenterol. 53, 1035–1047 (2018).
Howell, K. J. et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology 154, 585–598 (2018).
d’Aldebert, E. et al. Characterization of human colon organoids from inflammatory bowel disease patients. Front. Cell Dev. Biol. 8, 363 (2020).
Zhang, W. et al. Modeling microcephaly with cerebral organoids reveals a WDR62–CEP170–KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 10, 2612 (2019).
Raja, W. K. et al. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS ONE 11, e0161969 (2016).
Gonzalez, C. et al. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry 23, 2363–2374 (2018).
Chumarina, M. et al. Cellular alterations identified in pluripotent stem cell-derived midbrain spheroids generated from a female patient with progressive external ophthalmoplegia and parkinsonism who carries a novel variation (p.Q811R) in the POLG1 gene. Acta Neuropathol. Commun. 7, 208 (2019).
Buskin, A. et al. Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat. Commun. 9, 4234 (2018).
Parfitt, D. A. et al. Identification and correction of mechanisms underlying inherited blindness in human iPSC-derived optic cups. Cell Stem Cell 18, 769–781 (2016).
Li, G. et al. Generation and characterization of induced pluripotent stem cells and retinal organoids from a Leber’s congenital amaurosis patient with novel RPE65 mutations. Front. Mol. Neurosci. 12, 212 (2019).
Freedman, B. S. et al. Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. J. Am. Soc. Nephrol. 24, 1571–1586 (2013).
Cruz, N. M. et al. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 16, 1112–1119 (2017).
Akbari, S. et al. Robust, long-term culture of endoderm-derived hepatic organoids for disease modeling. Stem Cell Rep. 13, 627–641 (2019).
Guan, Y. et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2, e94954 (2017).
Ouchi, R. et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 30, 374–384.e6 (2019).
Rodansky, E. S., Johnson, L. A., Huang, S., Spence, J. R. & Higgins, P. D. R. Intestinal organoids: A model of intestinal fibrosis for evaluating anti-fibrotic drugs. Exp. Mol. Pathol. 98, 346–351 (2015).
Woo, D. et al. Enhancing a Wnt-telomere feedback loop restores intestinal stem cell function in a human organotypic model of dyskeratosis congenita. Cell Stem Cell 19, 397–405 (2016).
Fan, H., Demirci, U. & Chen, P. Emerging organoid models: leaping forward in cancer research. J. Hematol. Oncol. 12, 142 (2019).
Clevers, H. & Tuveson, D. A. Organoid models for cancer research. Annu. Rev. Cancer Biol. 3, 223–234 (2019).
Fujii, M. et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 18, 827–838 (2016).
Broutier, L. et al. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat. Med. 23, 1424–1435 (2017).
Cao, W. et al. Modeling liver cancer and therapy responsiveness using organoids derived from primary mouse liver tumors. Carcinogenesis 40, 145–154 (2019).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
Kim, M. et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 10, 3991 (2019).
Vlachogiannis, G. et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018).
Driehuis, E. et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl Acad. Sci. USA 116, 26580–26590 (2019).
Yan, H. H. N. et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell 23, 882–897.e11 (2018).
Bruna, A. et al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 167, 260–274.e22 (2016).
Zhou, T. et al. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 21, 274–283.e5 (2017).
Leite, S. B. et al. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials 78, 1–10 (2016).
Liu, F., Huang, J. & Liu, Z. Vincristine impairs microtubules and causes neurotoxicity in cerebral organoids. Neuroscience 404, 530–540 (2019).
Qian, X., Nguyen, H. N., Jacob, F., Song, H. & Ming, G. L. Using brain organoids to understand Zika virus-induced microcephaly. Development 144, 952–957 (2017).
Dutta, D., Heo, I. & Clevers, H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 23, 393–410 (2017).
Zhou, J. et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl Acad. Sci. USA 115, 6822–6827 (2018).
Hui, K. P. Y. et al. Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir. Med. 6, 846–854 (2018).
Monteil, V. et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181, 905–913.e7 (2020).
Lamers, M. M. et al. SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020).
Zhou, J. et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 26, 1077–1083 (2020).
Salahudeen, A. A. et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588, 670–675 (2020).
Han, Y. et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275 (2021).
Ettayebi, K. et al. Replication of human noroviruses in stem cell–derived human enteroids. Science 353, 1387–1393 (2016).
Zhang, Y.-G., Wu, S., Xia, Y. & Sun, J. Salmonella-infected crypt-derived intestinal organoid culture system for host–bacterial interactions. Physiol. Rep. 2, e12147 (2014).
Wilson, S. S., Tocchi, A., Holly, M. K., Parks, W. C. & Smith, J. G. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 8, 352–361 (2015).
Forbester, J. L. et al. Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 83, 2926–2934 (2015).
Finkbeiner, S. R. et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 3, e00159-12 (2012).
Porotto, M. et al. Authentic modeling of human respiratory virus infection in human pluripotent stem cell-derived lung organoids. mBio 10, e00723-19 (2019).
Ciancanelli, M. J. et al. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 348, 448–453 (2015).
Acknowledgements
The authors thank Stefano Davide Vianello for preparing the illustrations and his much-valued contributions editing the text, and thank Bilge Sen Elci and Antonius Chrisnandy for discussions and comments on the manuscript. M.P.L.’s work in the area of organoid biology and technology is supported by the Swiss National Science Foundation (#310030_179447), École Polytechnique Fédérale de Lausanne (EPFL), the European Union’s Horizon 2020 research and innovation programme (INTENS, #668294), the Personalized Health and Related Technologies initiative from the ETH Board, the National Center of Competence in Research (NCCR) Bio-Inspired Materials, the FreeNovation funding program of the Novartis Research Foundation, the Marie Skłodowska-Curie European Training Network ‘EUROoC’ (H2020-MSCA-ITN-2018) and the Vienna Science and Technology Fund (WWTF). The authors apologize to all researchers and colleagues whose work could not be considered.
Author information
Authors and Affiliations
Contributions
M.H. researched data for the article, made substantial contributions to discussions of the content and wrote the article. M.P.L. reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hofer, M., Lutolf, M.P. Engineering organoids. Nat Rev Mater 6, 402–420 (2021). https://doi.org/10.1038/s41578-021-00279-y
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41578-021-00279-y
This article is cited by
-
Organoids for disease modeling and treatment: state-of-the-art
Experimental Hematology & Oncology (2026)
-
Squidiff: predicting cellular development and responses to perturbations using a diffusion model
Nature Methods (2026)
-
Selecting the optimal cell migration assay: fundamentals and practical guidelines
Nature Methods (2026)
-
Disease exacerbation in human DMD MYOrganoids enables gene therapy evaluation and unveils persistence of fibrotic activity
npj Regenerative Medicine (2026)
-
Organoid-based systems for biomedical innovation: advances in disease modeling, drug screening, and precision medicine
Naunyn-Schmiedeberg's Archives of Pharmacology (2026)