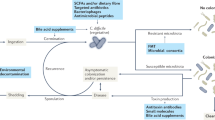
Abstract
Clostridioides difficile infection (CDI) continues to be a notable burden worldwide, both in terms of patient mortality and morbidity, and the economic costs associated with treatment, diagnosis and management. The epidemiology of C. difficile has changed markedly over the decades, with high CDI rates driven by clinical pressures exacerbated by the severe acute respiratory syndrome coronavirus 2 pandemic, antibiotic resistance and selective pressures caused by antimicrobial use. C. difficile is challenging to diagnose and treat as it forms spores and can persist asymptomatically within the gut. Some strains express multiple virulence factors, including adhesins and toxins. The gut microbiota is crucially important in CDI, as a healthy microbiota is resistant to colonization with C. difficile. Dysbiosis, often caused by antimicrobial exposure, enables C. difficile spores to germinate and produce toxin, causing symptoms that can range from mild diarrhoea to fulminant colitis and death. This Review describes changes in epidemiology and effects on diagnosis, discusses recent breakthroughs in the understanding of pathogenesis and antibiotic resistance and explores the role of microbiota dysbiosis in CDI and novel microbiota therapies in CDI treatment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lawson, P. A., Citron, D. M., Tyrrell, K. L. & Finegold, S. M. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 40, 95–99 (2016).
Kuehne, S. A. et al. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J. Infect. Dis. 209, 83–86 (2014). This article showcases the role of the different CDTs in causing infection.
van Prehn, J. et al. European society of clinical microbiology and infectious diseases: 2021 update on the treatment guidance document for Clostridioides difficile infection in adults. Clin. Microbiol. Infect. 27, S1–s21 (2021).
Mounsey, A., Lacy Smith, K., Reddy, V. C. & Nickolich, S. Clostridioides difficile infection: update on management. Am. Fam. Physician 101, 168–175 (2020).
Hall, I. C. & O’toole, E. Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. Am. J. Dis. Child. 49, 390–402 (1935).
George, W. L., Sutter, V. L., Goldstein, E. J., Ludwig, S. L. & Finegold, S. M. Aetiology of antimicrobial-agent-associated colitis. Lancet 1, 802–803 (1978).
Bartlett, J. G., Moon, N., Chang, T. W., Taylor, N. & Onderdonk, A. B. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 75, 778–782 (1978).
Tsigrelis, C. Recurrent Clostridioides difficile infection: recognition, management, prevention. Cleve. Clin. J. Med. 87, 347–359 (2020).
Guh, A. Y. et al. Characteristics of patients with initial Clostridioides difficile infection (CDI) that are associated with increased risk of multiple CDI recurrences. Open Forum Infect. Dis. 11, ofae127 (2024).
Olsen, M. A., Yan, Y., Reske, K. A., Zilberberg, M. D. & Dubberke, E. R. Recurrent Clostridium difficile infection is associated with increased mortality. Clin. Microbiol. Infect. 21, 164–170 (2015).
Olsen, M. A., Yan, Y., Reske, K. A., Zilberberg, M. & Dubberke, E. R. Impact of Clostridium difficile recurrence on hospital readmissions. Am. J. Infect. Control. 43, 318–322 (2015).
Tresman, R. & Goldenberg, S. D. Healthcare resource use and attributable cost of Clostridium difficile infection: a micro-costing analysis comparing first and recurrent episodes. J. Antimicrob. Chemother. 73, 2851–2855 (2018).
Fawley, W. N. et al. Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS ONE 10, e0118150 (2015).
Freeman, J. et al. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23, 529–549 (2010).
McDonald, L. C. et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious diseases society of America (IDSA) and Society for healthcare epidemiology of America (SHEA). Clin. Infect. Dis. 66, e1–e48 (2018).
Guh, A. Y. et al. Trends in U.S. burden of Clostridioides difficile infection and outcomes. N. Engl. J. Med. 382, 1320–1330 (2020).
Alshannaq, A. F. et al. Diverse sources and latent reservoirs of community-associated Clostridioides difficile infection. Clin. Infect. Dis. 80, 37–42 (2025).
Worley, J. et al. Genomic determination of relative risks for Clostridioides difficile infection from asymptomatic carriage in intensive care unit patients. Clin. Infect. Dis. 73, e1727–e1736 (2021).
Gilboa, M., Baharav, N., Melzer, E., Regev-Yochay, G. & Yahav, D. Screening for asymptomatic Clostridioides difficile carriage among hospitalized patients: a narrative review. Infect. Dis. Ther. 12, 2223–2240 (2023).
Caballero-Flores, G., Pickard, J. M. & Núñez, G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat. Rev. Microbiol. 21, 347–360 (2023).
Dingle, K. E. et al. Effects of control interventions on Clostridium difficile infection in England: an observational study. Lancet Infect. Dis. 17, 411–421 (2017).
Slimings, C. & Riley, T. V. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J. Antimicrob. Chemother. 69, 881–891 (2014).
Dingle, K. E. et al. Penicillin binding protein substitutions cooccur with fluoroquinolone resistance in epidemic lineages of multidrug-resistant Clostridioides difficile. mBio 14, e0024323 (2023).
Webb, B. J. et al. Antibiotic exposure and risk for hospital-associated Clostridioides difficile infection. Antimicrob. Agents Chemother. 64, e02169-19 (2020).
Johnson, S. et al. Clinical practice guideline by the infectious diseases society of America (IDSA) and society for healthcare epidemiology of America (SHEA): 2021 focused update guidelines on management of Clostridioides difficile infection in adults. Clin. Infect. Dis. 73, e1029–e1044 (2021).
Buckley, A. M., Moura, I. B. & Wilcox, M. H. The potential of microbiome replacement therapies for Clostridium difficile infection. Curr. Opin. Gastroenterol. 38, 1–6 (2022).
Wilcox, M. H. et al. Changing epidemiology of Clostridium difficile infection following the introduction of a national ribotyping-based surveillance scheme in England. Clin. Infect. Dis. 55, 1056–1063 (2012).
Agnew, E. et al. Impact of testing on Clostridioides difficile infection in hospitals across Europe: a mathematical model. Clin. Microbiol. Infect. 29, 796.e1–796.e6 (2023).
Tkalec, V. et al. Clostridioides difficile positivity rate and PCR ribotype distribution on retail potatoes in 12 European countries, January to June 2018. Eur. Surveill. 27, 2100417 (2022).
Rupnik, M. et al. Distribution of Clostridioides difficile ribotypes and sequence types across humans, animals and food in 13 European countries. Emerg. Microbes Infect. 13, 2427804 (2024).
Viprey, V. F. et al. A point-prevalence study on community and inpatient Clostridioides difficile infections (CDI): results from Combatting Bacterial Resistance in Europe CDI (COMBACTE-CDI), July to November 2018. Eur. Surveill. 27, 2100704 (2022). This article offers a detailed description of the rates of community and health-care-associated CDI across Europe, regardless of diagnosis.
Abdrabou, A. M. M. et al. Implementation of a Clostridioides difficile sentinel surveillance system in Germany: first insights for 2019-2021. Anaerobe 77, 102548 (2022).
Marujo, V. & Arvand, M. The largely unnoticed spread of Clostridioides difficile PCR ribotype 027 in Germany after 2010. Infect. Prev. Pract. 2, 100102 (2020).
Berger, F. K. et al. Hospital outbreak due to Clostridium difficile ribotype 018 (RT018) in Southern Germany. Int. J. Med. Microbiol. 309, 189–193 (2019).
Gateau, C. et al. Local outbreak of Clostridioides difficile PCR-ribotype 018 investigated by multi locus variable number tandem repeat analysis, whole genome multi locus sequence typing and core genome single nucleotide polymorphism typing. Anaerobe 60, 102087 (2019).
UK Health Security Agency. Clostridioides difficile Ribotyping Network (CDRN) for England and Northern Ireland, 2018 to 2023 (UKHSA, 2024).
UKHSA Advisory Board. Update on Preparedness for Infectious Diseases (UKHSA, 2024).
Kachrimanidou, M. et al. Predominance of Clostridioides difficile PCR ribotype 181 in northern Greece, 2016-2019. Anaerobe 76, 102601 (2022).
National Center for Emerging and Zoonotic Infectious Diseases. Emerging Infections Program, Healthcare-Associated Infections — Community Interface Surveillance Report, Clostridioides difficile Infection (CDI), 2021 (CDC, 2023).
Snydman, D. R. et al. A US-based national surveillance study for the susceptibility and epidemiology of Clostridioides difficile isolates with special reference to ridinilazole: 2020–2021. Antimicrob. Agents Chemother. 67, e0034923 (2023).
Canadian Nosocomial Infection Surveillance Program. Healthcare-associated infections and antimicrobial resistance in Canadian acute care hospitals, 2017–2021. Can. Commun. Dis. Rep. 49, 235–252 (2023).
Hong, S. et al. Laboratory-based surveillance of Clostridium difficile infection in Australian health care and community settings, 2013 to 2018. J. Clin. Microbiol. 58, e01552-20 (2020).
O’Grady, K. et al. Defining the phylogenetics and resistome of the major Clostridioides difficile ribotypes circulating in Australia. Microb. Genom. 10, 001232 (2024).
Angulo, F. J., Ghia, C., Fletcher, M. A., Ozbilgili, E. & Morales, G. D. C. The burden of Clostridioides difficile infections in South-East Asia and the Western Pacific: a narrative review. Anaerobe 86, 102821 (2024).
Brajerova, M., Zikova, J. & Krutova, M. Clostridioides difficile epidemiology in the Middle and the Far East. Anaerobe 74, 102542 (2022).
Acuña-Amador, L., Quesada-Gómez, C. & Rodríguez, C. Clostridioides difficile in Latin America: a comprehensive review of literature (1984-2021). Anaerobe 74, 102547 (2022).
Morales-Olvera, C. G. et al. Clostridioides Difficile in Latin America: an epidemiological overview. Curr. Microbiol. 80, 357 (2023).
Senoh, M. & Kato, H. Molecular epidemiology of endemic Clostridioides difficile infection in Japan. Anaerobe 74, 102510 (2022).
Bignardi, G. E., Hill, K., Berrington, A. & Settle, C. D. Two-stage algorithm for Clostridium difficile: glutamate-dehydrogenase-positive toxin-negative enzyme immunoassay results may require further testing. J. Hosp. Infect. 83, 347–349 (2013).
Planche, T. D. et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect. Dis. 13, 936–945 (2013). This article demonstrates how different diagnostic methods correlate with clinical outcomes and was instrumental in the development of optimized two-step diagnostic algorithms.
Iheagwara, C. C. et al. A rare case of polymerase chain reaction-negative severe Clostridioides difficile infection. Cureus 15, e50403 (2023).
Polage, C. R. et al. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern. Med. 175, 1792–1801 (2015).
Bartlett, J. G., Chang, T. W., Gurwith, M., Gorbach, S. L. & Onderdonk, A. B. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N. Engl. J. Med. 298, 531–534 (1978).
Chang, T. W., Gorbach, S. L. & Bartlett, J. B. Neutralization of Clostridium difficile toxin by Clostridium sordellii antitoxins. Infect. Immun. 22, 418–422 (1978).
Trubiano, J. A. et al. Australasian society of infectious diseases updated guidelines for the management of Clostridium difficile infection in adults and children in Australia and New Zealand. Intern. Med. J. 46, 479–493 (2016).
Watkin, S., Yongblah, F., Burton, J., Hartley, J. C. & Cloutman-Green, E. Clostridioides difficile detection and infection in children: are they just small adults? J. Med. Microbiol. 73, 001816 (2024).
Dalal, R. S. & Allegretti, J. R. Diagnosis and management of Clostridioides difficile infection in patients with inflammatory bowel disease. Curr. Opin. Gastroenterol. 37, 336–343 (2021).
Crobach, M. J. et al. European society of clinical microbiology and infectious diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 22, S63–81 (2016).
Viprey, V. F. et al. European survey on the current surveillance practices, management guidelines, treatment pathways and heterogeneity of testing of Clostridioides difficile, 2018–2019: results from the Combatting Bacterial Resistance in Europe CDI (COMBACTE-CDI). J. Hosp. Infect. 131, 213–220 (2023).
Planche, T. et al. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect. Dis. 8, 777–784 (2008).
Tichota-Lee, J., Jaqua-Stewart, M. J., Benfield, D., Simmons, J. L. & Jaqua, R. A. Effect of age on the sensitivity of cell cultures to Clostridium difficile toxin. Diagn. Microbiol. Infect. Dis. 8, 203–214 (1987).
Lyerly, D. M., Sullivan, N. M. & Wilkins, T. D. Enzyme-linked immunosorbent assay for Clostridium difficile toxin A. J. Clin. Microbiol. 17, 72–78 (1983).
Borriello, S. P. et al. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60, 4192–4199 (1992).
Eastwood, K., Else, P., Charlett, A. & Wilcox, M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J. Clin. Microbiol. 47, 3211–3217 (2009).
Tenover, F. C. et al. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J. Clin. Microbiol. 48, 3719–3724 (2010).
Mansfield, M. J. et al. Phylogenomics of 8,839 Clostridioides difficile genomes reveals recombination-driven evolution and diversification of toxin A and B. PLoS Pathog. 16, e1009181 (2020).
Ramírez-Vargas, G. et al. Novel clade C-I Clostridium difficile strains escape diagnostic tests, differ in pathogenicity potential and carry toxins on extrachromosomal elements. Sci. Rep. 8, 13951 (2018).
Shen, E. et al. Subtyping analysis reveals new variants and accelerated evolution of Clostridioides difficile toxin B. Commun. Biol. 3, 347 (2020).
Landry, M. L. et al. High agreement between an ultrasensitive Clostridioides difficile toxin assay and a C. difficile laboratory algorithm utilizing GDH-and-toxin enzyme immunoassays and cytotoxin testing. J. Clin. Microbiol. 58, e01629-19 (2020).
Sandlund, J. et al. Ultrasensitive detection of Clostridioides difficile toxins A and B by use of automated single-molecule counting technology. J. Clin. Microbiol. 56, e00908-18 (2018).
Banz, A. et al. Sensitivity of single-molecule array assays for detection of Clostridium difficile toxins in comparison to conventional laboratory testing algorithms. J. Clin. Microbiol. 56, e00452-18 (2018).
Shim, J. K., Johnson, S., Samore, M. H., Bliss, D. Z. & Gerding, D. N. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 351, 633–636 (1998).
Bouza, E. et al. ‘Second-look’ cytotoxicity: an evaluation of culture plus cytotoxin assay of Clostridium difficile isolates in the laboratory diagnosis of CDAD. J. Hosp. Infect. 48, 233–237 (2001).
Anderson, B. M., Anderson, C. D., Van Tassell, R. L., Lyerly, D. M. & Wilkins, T. D. Purification and characterization of Clostridium difficile glutamate dehydrogenase. Arch. Biochem. Biophys. 300, 483–488 (1993).
Shetty, N., Wren, M. W. & Coen, P. G. The role of glutamate dehydrogenase for the detection of Clostridium difficile in faecal samples: a meta-analysis. J. Hosp. Infect. 77, 1–6 (2011).
Avni, T. et al. Molecular-based diagnosis of Clostridium difficile infection is associated with reduced mortality. Eur. J. Clin. Microbiol. Infect. Dis. 37, 1137–1142 (2018).
Longtin, Y. et al. Impact of the type of diagnostic assay on Clostridium difficile infection and complication rates in a mandatory reporting program. Clin. Infect. Dis. 56, 67–73 (2013).
Prosty, C. et al. Clinical outcomes and management of NAAT-positive/toxin-negative Clostridioides difficile infection: a systematic review and meta-analysis. Clin. Infect. Dis. 78, 430–438 (2024).
Goret, J. et al. Impact of the introduction of a nucleic acid amplification test for Clostridium difficile diagnosis on stool rejection policies. Gut Pathog. 10, 19 (2018).
Yen, C. et al. Reducing Clostridium difficile colitis rates via cost-saving diagnostic stewardship. Infect. Control. Hosp. Epidemiol. 39, 734–736 (2018).
Davies, K. A. et al. Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID). Lancet Infect. Dis. 14, 1208–1219 (2014).
Davies, K. et al. Variability in testing policies and impact on reported Clostridium difficile infection rates: results from the pilot longitudinal European Clostridium difficile infection diagnosis surveillance study (LuCID). Eur. J. Clin. Microbiol. Infect. Dis. 35, 1949–1956 (2016).
Lessa, F. C. et al. Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 825–834 (2015).
Davies, K. A., Planche, T. & Wilcox, M. H. The predictive value of quantitative nucleic acid amplification detection of Clostridium difficile toxin gene for faecal sample toxin status and patient outcome. PLoS ONE 13, e0205941 (2018).
Goldenberg, S. D. & French, G. L. Lack of association of tcdC type and binary toxin status with disease severity and outcome in toxigenic Clostridium difficile. J. Infect. 62, 355–362 (2011).
Pollock, N. R. Ultrasensitive detection and quantification of toxins for optimized diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 54, 259–264 (2016).
Pollock, N. R. et al. Comparison of Clostridioides difficile stool toxin concentrations in adults with symptomatic infection and asymptomatic carriage using an ultrasensitive quantitative immunoassay. Clin. Infect. Dis. 68, 78–86 (2019).
Ford, C. D., Lopansri, B. K., Hunter, B. D., Asch, J. & Hoda, D. Multiplexed gastrointestinal PCR panels for the evaluation of diarrhea in HCT recipients. Transpl. Cell Ther. 30, 814.e1–814.e7 (2024).
Langhorst, J. & Boone, J. Fecal lactoferrin as a noninvasive biomarker in inflammatory bowel diseases. Drugs Today 48, 149–161 (2012).
Barbut, F. et al. Faecal lactoferrin and calprotectin in patients with Clostridium difficile infection: a case-control study. Eur. J. Clin. Microbiol. Infect. Dis. 36, 2423–2430 (2017).
Swale, A. et al. Calprotectin and lactoferrin faecal levels in patients with Clostridium difficile infection (CDI): a prospective cohort study. PLoS ONE 9, e106118 (2014).
Lee, S. H. et al. Fecal calprotectin predicts complete mucosal healing and better correlates with the ulcerative colitis endoscopic index of severity than with the Mayo endoscopic subscore in patients with ulcerative colitis. BMC Gastroenterol. 17, 110 (2017).
O’Connor, J. R., Johnson, S. & Gerding, D. N. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology 136, 1913–1924 (2009).
Wickramage, I., Spigaglia, P. & Sun, X. Mechanisms of antibiotic resistance of Clostridioides difficile. J. Antimicrob. Chemother. 76, 3077–3090 (2021).
Vernon, J. J., Wilcox, M. H. & Freeman, J. Antimicrobial resistance progression in the United Kingdom: a temporal comparison of Clostridioides difficile antimicrobial susceptibilities. Anaerobe 70, 102385 (2021).
National Insitute for Health and Care Excellence. Scenario: management of antibiotic associated diarrhoea. NICE https://cks.nice.org.uk/topics/diarrhoea-antibiotic-associated/management/diarrhoea-antibiotic-associated/#suspected-or-confirmed-clostridiodes-difficile-infections (2023).
Gonzales-Luna, A. J. et al. Reduced susceptibility to metronidazole is associated with initial clinical failure in Clostridioides difficile infection. Open Forum Infect. Dis. 8, ofab365 (2021).
Freeman, J. et al. The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011–2014. Clin. Microbiol. Infect. 24, 724–731 (2018).
Goudarzi, M. et al. Antimicrobial susceptibility of Clostridium difficile clinical isolates in Iran. Iran. Red. Crescent Med. J. 15, 704–711 (2013).
European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 15.0 (EUCAST, 2025).
Peláez, T. et al. Metronidazole resistance in Clostridium difficile is heterogeneous. J. Clin. Microbiol. 46, 3028–3032 (2008).
Moura, I., Spigaglia, P., Barbanti, F. & Mastrantonio, P. Analysis of metronidazole susceptibility in different Clostridium difficile PCR ribotypes. J. Antimicrob. Chemother. 68, 362–365 (2013).
Baines, S. D. et al. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J. Antimicrob. Chemother. 62, 1046–1052 (2008).
Boekhoud, I. M. et al. Plasmid-mediated metronidazole resistance in Clostridioides difficile. Nat. Commun. 11, 598 (2020). This article describes a novel plasmid linked to metronidazole resistance in C. difficile.
Imwattana, K., Rodríguez, C., Riley, T. V. & Knight, D. R. A species-wide genetic atlas of antimicrobial resistance in Clostridioides difficile. Microb. Genom. 7, 000696 (2021).
Boekhoud, I. M. et al. Haem is crucial for medium-dependent metronidazole resistance in clinical isolates of Clostridioides difficile. J. Antimicrob. Chemother. 76, 1731–1740 (2021).
Olaitan, A. O. et al. Decoding a cryptic mechanism of metronidazole resistance among globally disseminated fluoroquinolone-resistant Clostridioides difficile. Nat. Commun. 14, 4130 (2023).
Ammam, F. et al. The functional vanGCd cluster of Clostridium difficile does not confer vancomycin resistance. Mol. Microbiol. 89, 612–625 (2013).
Eubank, T. A., Gonzales-Luna, A. J., Hurdle, J. G. & Garey, K. W. Genetic mechanisms of vancomycin resistance in Clostridioides difficile: a systematic review. Antibiotics 11, 258 (2022).
Shen, W. J. et al. Constitutive expression of the cryptic vanGCd operon promotes vancomycin resistance in Clostridioides difficile clinical isolates. J. Antimicrob. Chemother. 75, 859–867 (2020).
Buddle, J. E. et al. Identification of pathways to high-level vancomycin resistance in Clostridioides difficile that incur high fitness costs in key pathogenicity traits. PLoS Biol. 22, e3002741 (2024).
Tsvetkova, K., Marvaud, J. C. & Lambert, T. Analysis of the mobilization functions of the vancomycin resistance transposon Tn1549, a member of a new family of conjugative elements. J. Bacteriol. 192, 702–713 (2010).
Wu, Y. et al. Independent microevolution mediated by mobile genetic elements of individual Clostridium difficile isolates from clade 4 revealed by whole-genome sequencing. mSystems 4, e00252-18 (2019).
Pu, M. et al. Plasmid acquisition alters vancomycin susceptibility in Clostridioides difficile. Gastroenterology 160, 941–945.e8 (2021).
Saldanha, G. Z. et al. Genetic relatedness, virulence factors and antimicrobial resistance of C. difficile strains from hospitalized patients in a multicentric study in Brazil. J. Glob. Antimicrob. Resist. 22, 117–121 (2020).
Eubank, T. A., Dureja, C., Garey, K. W., Hurdle, J. G. & Gonzales-Luna, A. J. Reduced vancomycin susceptibility in Clostridioides difficile is associated with lower rates of initial cure and sustained clinical response. Clin. Infect. Dis. 79, 15–21 (2024).
Goldstein, E. J. et al. Comparative susceptibilities to fidaxomicin (OPT-80) of isolates collected at baseline, recurrence, and failure from patients in two phase III trials of fidaxomicin against Clostridium difficile infection. Antimicrob. Agents Chemother. 55, 5194–5199 (2011).
Schwanbeck, J. et al. Characterization of a clinical Clostridioides difficile isolate with markedly reduced fidaxomicin susceptibility and a V1143D mutation in rpoB. J. Antimicrob. Chemother. 74, 6–10 (2019).
Marchandin, H. et al. In vivo emergence of a still uncommon resistance to fidaxomicin in the urgent antimicrobial resistance threat Clostridioides difficile. J. Antimicrob. Chemother. 78, 1992–1999 (2023).
Aoki, K., Takeda, S., Miki, T., Ishii, Y. & Tateda, K. Antimicrobial susceptibility and molecular characterisation using whole-genome sequencing of Clostridioides difficile collected in 82 hospitals in Japan between 2014 and 2016. Antimicrob. Agents Chemother. 63, e01259-19 (2019).
Redmond, S. N. et al. Emergence and spread of Clostridioides difficile isolates with reduced fidaxomicin susceptibility in an acute care hospital. Clin. Infect. Dis. 80, 984–991 (2025).
Toth, M., Stewart, N. K., Smith, C. & Vakulenko, S. B. Intrinsic class D β-lactamases of Clostridium difficile. mBio 9, e01803-18 (2018).
Isidro, J. et al. Imipenem resistance in Clostridium difficile ribotype 017, Portugal. Emerg. Infect. Dis. 24, 741–745 (2018).
Sacco, M. D. et al. A unique class of Zn2+-binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile. Nat. Commun. 13, 4370 (2022).
Curry, S. R. et al. High frequency of rifampin resistance identified in an epidemic Clostridium difficile clone from a large teaching hospital. Clin. Infect. Dis. 48, 425–429 (2009).
Carman, R. J., Boone, J. H., Grover, H., Wickham, K. N. & Chen, L. In vivo selection of rifamycin-resistant Clostridium difficile during rifaximin therapy. Antimicrob. Agents Chemother. 56, 6019–6020 (2012).
Dang, U. T. et al. Rifamycin resistance in Clostridium difficile is generally associated with a low fitness burden. Antimicrob. Agents Chemother. 60, 5604–5607 (2016).
Corver, J. et al. Analysis of a Clostridium difficile PCR ribotype 078 100 kilobase island reveals the presence of a novel transposon, Tn6164. BMC Microbiol. 12, 130 (2012).
Spigaglia, P., Barbanti, F. & Mastrantonio, P. Tetracycline resistance gene tet(W) in the pathogenic bacterium Clostridium difficile. Antimicrob. Agents Chemother. 52, 770–773 (2008).
Deakin, L. J. et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect. Immun. 80, 2704–2711 (2012).
Péchiné, S., Denève-Larrazet, C. & Collignon, A. Clostridium difficile adhesins. Methods Mol. Biol. 1476, 91–101 (2016).
Papatheodorou, P., Minton, N. P., Aktories, K. & Barth, H. An updated view on the cellular uptake and mode-of-action of Clostridioides difficile toxins. Adv. Exp. Med. Biol. 1435, 219–247 (2024).
Buddle, J. E. & Fagan, R. P. Pathogenicity and virulence of Clostridioides difficile. Virulence 14, 2150452 (2023).
Martins, D. et al. CD25890, a conserved protein that modulates sporulation initiation in Clostridioides difficile. Sci. Rep. 11, 7887 (2021).
DiCandia, M. A. et al. Identification of functional Spo0A residues critical for sporulation in Clostridioides difficile. J. Mol. Biol. 434, 167641 (2022).
Lee, C. D. et al. Genetic mechanisms governing sporulation initiation in Clostridioides difficile. Curr. Opin. Microbiol. 66, 32–38 (2022).
DiCandia, M. A. et al. A conserved switch controls virulence, sporulation, and motility in C. difficile. PLoS Pathog. 20, e1012224 (2024).
Edwards, A. N., Krall, E. G. & McBride, S. M. Strain-dependent RstA regulation of Clostridioides difficile toxin production and sporulation. J. Bacteriol. 202, e00586-19 (2020).
Paredes-Sabja, D., Shen, A. & Sorg, J. A. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 22, 406–416 (2014).
Montes-Bravo, N. et al. Role of the spore coat proteins CotA and CotB, and the spore surface protein CDIF630_02480, on the surface distribution of exosporium proteins in Clostridioides difficile 630 spores. Microorganisms 10, 1918 (2022).
Pizarro-Guajardo, M., Calderón-Romero, P., Castro-Córdova, P., Mora-Uribe, P. & Paredes-Sabja, D. Ultrastructural variability of the exosporium layer of Clostridium difficile spores. Appl. Environ. Microbiol. 82, 2202–2209 (2016).
Pizarro-Guajardo, M. et al. Characterization of the collagen-like exosporium protein, BclA1, of Clostridium difficile spores. Anaerobe 25, 18–30 (2014).
Sorg, J. A. & Sonenshein, A. L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512 (2008).
Kochan, T. J. et al. Intestinal calcium and bile salts facilitate germination of Clostridium difficile spores. PLoS Pathog. 13, e1006443 (2017).
Francis, M. B., Allen, C. A., Shrestha, R. & Sorg, J. A. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog. 9, e1003356 (2013).
Shrestha, R., Cochran, A. M. & Sorg, J. A. The requirement for co-germinants during Clostridium difficile spore germination is influenced by mutations in yabG and cspA. PLoS Pathog. 15, e1007681 (2019).
Kevorkian, Y. & Shen, A. Revisiting the role of Csp family proteins in regulating Clostridium difficile spore germination. J. Bacteriol. 199, e00266-17 (2017).
Rohlfing, A. E. et al. The CspC pseudoprotease regulates germination of Clostridioides difficile spores in response to multiple environmental signals. PLoS Genet. 15, e1008224 (2019).
Ribis, J. W. et al. Single-spore germination analyses reveal that calcium released during Clostridioides difficile germination functions in a feedforward loop. mSphere 8, e0000523 (2023).
Castro-Córdova, P. et al. Redistribution of the novel Clostridioides difficile spore adherence receptor E-cadherin by TcdA and TcdB increases spore binding to adherens junctions. Infect. Immun. 91, e0047622 (2023).
Normington, C. et al. Biofilms harbour Clostridioides difficile, serving as a reservoir for recurrent infection. npj Biofilms Microbiomes 7, 16 (2021). This article demonstrates the role of. C. difficile spores in recurrent CDI and pathogen persistence in the large intestine.
Ðapa, T. et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 195, 545–555 (2013).
Donelli, G., Vuotto, C., Cardines, R. & Mastrantonio, P. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol. Med. Microbiol. 65, 318–325 (2012).
Semenyuk, E. G. et al. Analysis of bacterial communities during Clostridium difficile infection in the mouse. Infect. Immun. 83, 4383–4391 (2015).
Smith, A. B. et al. Enterococci enhance Clostridioides difficile pathogenesis. Nature 611, 780–786 (2022).
Dubois, T. et al. A microbiota-generated bile salt induces biofilm formation in Clostridium difficile. npj Biofilms Microbiomes 5, 14 (2019).
Kang, J. D. et al. Bile acid 7α-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: role of secondary bile acids. Cell Chem. Biol. 26, 27–34.e24 (2019).
Baban, S. T. et al. The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS ONE 8, e73026 (2013).
McKee, R. W., Aleksanyan, N., Garrett, E. M. & Tamayo, R. Type IV Pili promote Clostridium difficile adherence and persistence in a mouse model of infection. Infect. Immun. 86, e00943-17 (2018).
Waligora, A. J. et al. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect. Immun. 69, 2144–2153 (2001).
Fagan, R. P. & Fairweather, N. F. Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222 (2014).
Kirk, J. A., Banerji, O. & Fagan, R. P. Characteristics of the Clostridium difficile cell envelope and its importance in therapeutics. Microb. Biotechnol. 10, 76–90 (2017).
Tasteyre, A., Barc, M. C., Collignon, A., Boureau, H. & Karjalainen, T. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69, 7937–7940 (2001).
Merrigan, M. M. et al. Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS ONE 8, e78404 (2013).
Calabi, E., Calabi, F., Phillips, A. D. & Fairweather, N. F. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70, 5770–5778 (2002).
Kirk, J. A. et al. New class of precision antimicrobials redefines role of Clostridium difficile S-layer in virulence and viability. Sci. Transl Med. 9, eaah6813 (2017).
Lanzoni-Mangutchi, P. et al. Structure and assembly of the S-layer in C. difficile. Nat. Commun. 13, 970 (2022).
Wang, S. et al. Revealing roles of S-layer protein (SlpA) in Clostridioides difficile pathogenicity by generating the first slpA gene deletion mutant. Microbiol. Spectr. 12, e0400523 (2024).
Kirby, J. M. et al. Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J. Biol. Chem. 284, 34666–34673 (2009).
Péchiné, S. et al. Immunological properties of surface proteins of Clostridium difficile. J. Med. Microbiol. 54, 193–196 (2005).
Wright, A., Drudy, D., Kyne, L., Brown, K. & Fairweather, N. F. Immunoreactive cell wall proteins of Clostridium difficile identified by human sera. J. Med. Microbiol. 57, 750–756 (2008).
Zhou, Q. et al. The cwp66 gene affects cell adhesion, stress tolerance, and antibiotic resistance in Clostridioides difficile. Microbiol. Spectr. 10, e0270421 (2022).
Schöttelndreier, D., Langejürgen, A., Lindner, R. & Genth, H. Low density lipoprotein receptor-related protein-1 (LRP1) is involved in the uptake of Clostridioides difficile toxin A and serves as an internalizing receptor. Front. Cell Infect. Microbiol. 10, 565465 (2020).
Tao, L. et al. Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538, 350–355 (2016).
Gupta, P. et al. Functional defects in Clostridium difficile TcdB toxin uptake identify CSPG4 receptor-binding determinants. J. Biol. Chem. 292, 17290–17301 (2017).
LaFrance, M. E. et al. Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc. Natl Acad. Sci. USA 112, 7073–7078 (2015).
Luo, J. et al. TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile. Cell 185, 980–994.e15 (2022).
Manion, J. et al. C. difficile intoxicates neurons and pericytes to drive neurogenic inflammation. Nature 622, 611–618 (2023).
Lyras, D. et al. Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179 (2009).
Mileto, S. J. et al. Clostridioides difficile infection damages colonic stem cells via TcdB, impairing epithelial repair and recovery from disease. Proc. Natl Acad. Sci. USA 117, 8064–8073 (2020).
Lv, X. et al. De novo design of mini-protein binders broadly neutralizing Clostridioides difficile toxin B variants. Nat. Commun. 15, 8521 (2024).
Eckert, C. et al. Prevalence and pathogenicity of binary toxin-positive Clostridium difficile strains that do not produce toxins A and B. New Microbes New Infect. 3, 12–17 (2015).
Gerding, D. N., Johnson, S., Rupnik, M. & Aktories, K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes 5, 15–27 (2014).
Lyon, S. A., Hutton, M. L., Rood, J. I., Cheung, J. K. & Lyras, D. CdtR regulates TcdA and TcdB production in Clostridium difficile. PLoS Pathog. 12, e1005758 (2016).
Dong, Q. et al. Virulence and genomic diversity among clinical isolates of ST1 (BI/NAP1/027) Clostridioides difficile. Cell Rep. 42, 112861 (2023).
Meza-Torres, J. et al. Clostridioides difficile binary toxin CDT induces biofilm-like persisting microcolonies. Gut Microbes 17, 2444411 (2025).
Nabukhotna, K. et al. Purified CDT toxins and a clean deletion within the CDT locus provide novel insights into the contribution of binary toxin in cellular inflammation and Clostridioides difficile infection. PLoS Pathog. 20, e1012568 (2024).
Simpson, M. et al. Clostridioides difficile binary toxin binding component increases virulence in a Hamster model. Open Forum Infect. Dis. 10, ofad040 (2023).
Nhu, N. T. Q. et al. Flagellar switch inverted repeats impact heterogeneity in flagellar gene expression and thus C. difficile RT027/MLST1 virulence. Cell Rep. 44, 115830 (2025).
Trzilova, D., Anjuwon-Foster, B. R., Torres Rivera, D. & Tamayo, R. Rho factor mediates flagellum and toxin phase variation and impacts virulence in Clostridioides difficile. PLoS Pathog. 16, e1008708 (2020).
Warren Norris, M. A. H., Plaskon, D. M. & Tamayo, R. Phase variation of flagella and toxins in Clostridioides difficile is mediated by selective Rho-dependent termination. J. Mol. Biol. 436, 168456 (2024).
Trzilova, D., Warren, M. A. H., Gadda, N. C., Williams, C. L. & Tamayo, R. Flagellum and toxin phase variation impacts intestinal colonization and disease development in a mouse model of Clostridioides difficile infection. Gut Microbes 14, 2038854 (2022).
Kelly, C. P. & Kyne, L. The host immune response to Clostridium difficile. J. Med. Microbiol. 60, 1070–1079 (2011).
Gupta, S. B. et al. Antibodies to toxin B are protective against Clostridium difficile infection recurrence. Clin. Infect. Dis. 63, 730–734 (2016).
Rigo, I. et al. The impact of existing total anti-toxin B IgG immunity in outcomes of recurrent Clostridioides difficile infection. Anaerobe 87, 102842 (2024).
Janoir, C. Virulence factors of Clostridium difficile and their role during infection. Anaerobe 37, 13–24 (2016).
Di Bella, S. et al. Clostridioides difficile infection: history, epidemiology, risk factors, prevention, clinical manifestations, treatment, and future options. Clin. Microbiol. Rev. 37, e0013523 (2024).
Abt, M. C. et al. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe 18, 27–37 (2015).
Saleh, M. M. & Petri, W. A. Jr. Type 3 immunity during Clostridioides difficile infection: too much of a good thing? Infect. Immun. 88, e00306–19 (2019).
Pruss, K. M. & Sonnenburg, J. L. C. difficile exploits a host metabolite produced during toxin-mediated disease. Nature 593, 261–265 (2021).
Pi, H. et al. Clostridioides difficile ferrosome organelles combat nutritional immunity. Nature 623, 1009–1016 (2023).
Cowardin, C. A. et al. The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat. Microbiol. 1, 16108 (2016).
Berg, G. et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome 8, 103 (2020).
Ivanov, I. I. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Cash, H. L., Whitham, C. V., Behrendt, C. L. & Hooper, L. V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Cario, E., Gerken, G. & Podolsky, D. K. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 132, 1359–1374 (2007).
De Vadder, F. et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156, 84–96 (2014).
Hugon, P. et al. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis. 15, 1211–1219 (2015).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Shin, J. H. et al. Bacteroides and related species: the keystone taxa of the human gut microbiota. Anaerobe 85, 102819 (2024).
Sorbara, M. T. & Pamer, E. G. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol. 12, 1–9 (2019).
Kommineni, S. et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–722 (2015).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). This study demonstrates a specific bile-acid mediated mechanistic pathway by which C. scindens contributes to gut microbiota colonization resistance against. C. difficile.
Girinathan, B. P. et al. In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe 29, 1693–1708.e7 (2021).
Pruss, K. M. et al. Oxidative ornithine metabolism supports non-inflammatory C. difficile colonization. Nat. Metab. 4, 19–28 (2022).
Arrieta-Ortiz, M. L. et al. Predictive regulatory and metabolic network models for systems analysis of Clostridioides difficile. Cell Host Microbe 29, 1709–1723.e5 (2021).
Theriot, C. M. & Young, V. B. Microbial and metabolic interactions between the gastrointestinal tract and Clostridium difficile infection. Gut Microbes 5, 86–95 (2014).
Berkell, M. et al. Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat. Commun. 12, 2241 (2021).
Vijay, A. & Valdes, A. M. Role of the gut microbiome in chronic diseases: a narrative review. Eur. J. Clin. Nutr. 76, 489–501 (2022).
von Schwartzenberg, R. J. et al. Caloric restriction disrupts the microbiota and colonization resistance. Nature 595, 272–277 (2021).
Denny, J. E., Flores, J. N., Mdluli, N. V. & Abt, M. C. Standard mouse diets lead to differences in severity in infectious and non-infectious colitis. mBio 16, e0330224 (2025).
Seekatz, A. M. et al. Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe 53, 64–73 (2018).
Seekatz, A. M. et al. Recovery of the gut microbiome following fecal microbiota transplantation. mBio 5, e00893-14 (2014).
Gulati, M., Singh, S. K., Corrie, L., Kaur, I. P. & Chandwani, L. Delivery routes for faecal microbiota transplants: available, anticipated and aspired. Pharmacol. Res. 159, 104954 (2020).
Tariq, R., Pardi, D. S., Bartlett, M. G. & Khanna, S. Low cure rates in controlled trials of fecal microbiota transplantation for recurrent Clostridium difficile infection: a systematic review and meta-analysis. Clin. Infect. Dis. 68, 1351–1358 (2019).
Littmann, E. R. et al. Host immunity modulates the efficacy of microbiota transplantation for treatment of Clostridioides difficile infection. Nat. Commun. 12, 755 (2021).
Dehlholm-Lambertsen, E. et al. Cost savings following faecal microbiota transplantation for recurrent Clostridium difficile infection. Ther. Adv. Gastroenterol. 12, 1756284819843002 (2019).
Zellmer, C. et al. Shiga toxin-producing Escherichia coli transmission via fecal microbiota transplant. Clin. Infect. Dis. 72, e876–e880 (2021).
DeFilipp, Z. et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050 (2019).
Theriot, C. M., Bowman, A. A. & Young, V. B. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1, e00045-15 (2016).
Lee, C. et al. Safety of fecal microbiota, live-jslm (REBYOTA) in individuals with recurrent Clostridioides difficile infection: data from five prospective clinical trials. Ther. Adv. Gastroenterol. 16, 17562848231174277 (2023).
Khanna, S. et al. Efficacy and safety of RBX2660 in PUNCH CD3, a phase III, randomized, double-blind, placebo-controlled trial with a bayesian primary analysis for the prevention of recurrent Clostridioides difficile infection. Drugs 82, 1527–1538 (2022). This phase III, double-blind, randomized, placebo-controlled clinical trial demonstrates that RBX2660 is a safe and effective treatment to reduce recurrent CDI following standard-of-care antibiotic therapy, with a sustained response.
Feuerstadt, P. et al. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. N. Engl. J. Med. 386, 220–229 (2022). This phase III, double-blind, randomized, placebo-controlled clinical trial demonstrates that administration of SER-109 following standard-of-care antibiotic therapy was superior in preventing recurrence of symptoms than placebo.
Cohen, S. H. et al. Extended follow-up of microbiome therapeutic SER-109 through 24 weeks for recurrent Clostridioides difficile infection in a randomized clinical trial. JAMA 328, 2062–2064 (2022).
Wilcox, M. H. et al. Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N. Engl. J. Med. 376, 305–317 (2017).
Tkaczyk, C. et al. Anti-toxin B neutralizing monoclonal antibody AZD5148 provides protection in a Clostridioides difficile gnotobiotic piglet model. Open Forum Infect. Dis. 12, ofae631.1244 (2025).
Hutton, M. L. et al. Bovine antibodies targeting primary and recurrent Clostridium difficile disease are a potent antibiotic alternative. Sci. Rep. 7, 3665 (2017).
Gerding, D. N. et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: a randomized clinical trial. JAMA 313, 1719–1727 (2015).
Johnson, S. et al. Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: results from two multinational, randomized, controlled trials. Clin. Infect. Dis. 59, 345–354 (2014).
Daley, P. et al. Surotomycin versus vancomycin in adults with Clostridium difficile infection: primary clinical outcomes from the second pivotal, randomized, double-blind, phase 3 trial. J. Antimicrob. Chemother. 72, 3462–3470 (2017).
Gerding, D. N. et al. Cadazolid for the treatment of Clostridium difficile infection: results of two double-blind, placebo-controlled, non-inferiority, randomised phase 3 trials. Lancet Infect. Dis. 19, 265–274 (2019).
Okhuysen, P. C. et al. A randomized, double-blind, phase 3 safety and efficacy study of ridinilazole versus vancomycin for treatment of Clostridioides difficile infection: clinical outcomes with microbiome and metabolome correlates of response. Clin. Infect. Dis. 78, 1462–1472 (2024).
Fujimoto, K. & Uematsu, S. Phage therapy for Clostridioides difficile infection. Front. Immunol. 13, 1057892 (2022).
Riley, T. V., Lyras, D. & Douce, G. R. Status of vaccine research and development for Clostridium difficile. Vaccine 37, 7300–7306 (2019).
Donskey, C. J. et al. CLOVER (CLOstridium difficile Vaccine Efficacy tRial) study: a phase 3, randomized trial investigating the efficacy and safety of a detoxified toxin A/B vaccine in adults 50 years and older at increased risk of Clostridioides difficile infection. Clin. Infect. Dis. 79, 1503–1511 (2024).
Alameh, M. G. et al. A multivalent mRNA-LNP vaccine protects against Clostridioides difficile infection. Science 386, 69–75 (2024).
Wasels, F., Spigaglia, P., Barbanti, F. & Mastrantonio, P. Clostridium difficile erm(B)-containing elements and the burden on the in vitro fitness. J. Med. Microbiol. 62, 1461–1467 (2013).
Stojković, V. et al. cfr(B), cfr(C), and a new cfr-like gene, cfr(E), in Clostridium difficile strains recovered across Latin America. Antimicrob. Agents Chemother. 64, e01074-19 (2019).
Ackermann, G. et al. Resistance to moxifloxacin in toxigenic Clostridium difficile isolates is associated with mutations in gyrA. Antimicrob. Agents Chemother. 45, 2348–2353 (2001).
Drudy, D. et al. High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J. Antimicrob. Chemother. 58, 1264–1267 (2006).
Sandhu, B. K., Edwards, A. N., Anderson, S. E., Woods, E. C. & McBride, S. M. Regulation and anaerobic function of the Clostridioides difficile β-lactamase. Antimicrob. Agents Chemother. 64, e01496-19 (2019).
Lyras, D. et al. Chloramphenicol resistance in Clostridium difficile is encoded on Tn4453 transposons that are closely related to Tn4451 from Clostridium perfringens. Antimicrob. Agents Chemother. 42, 1563–1567 (1998).
Persson, S. et al. Sentinel surveillance and epidemiology of Clostridioides difficile in Denmark, 2016 to 2019. Eur. Surveill. 27, 2200244 (2022).
European Centre for Disease Prevention and Control. Clostridioides difficile Infections — Annual Epidemiological Report for 2018–2020 (ECDC, 2024).
Azimirad, M. et al. Clostridioides difficile ribotypes 001 and 126 were predominant in Tehran healthcare settings from 2004 to 2018: a 14-year-long cross-sectional study. Emerg. Microbes Infect. 9, 1432–1443 (2020).
Kullin, B., Abratt, V. R., Reid, S. J. & Riley, T. V. Clostridioides difficile infection in Africa: a narrative review. Anaerobe 74, 102549 (2022).
Griffiths, D. et al. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 48, 770–778 (2010).
Huber, C. A., Foster, N. F., Riley, T. V. & Paterson, D. L. Challenges for standardization of Clostridium difficile typing methods. J. Clin. Microbiol. 51, 2810–2814 (2013).
European Centre for Disease Prevention and Control. Study Protocol for a Survey of Whole Genome Sequencing of Clostridioides difficile Isolates from Tertiary Acute Care Hospitals, EU/EEA, 2022–2023 (ECDC, 2024).
Baktash, A. et al. Comparison of whole-genome sequence-based methods and PCR ribotyping for subtyping of Clostridioides difficile. J. Clin. Microbiol. 60, e0173721 (2022).
Knight, D. R. et al. Major genetic discontinuity and novel toxigenic species in Clostridioides difficile taxonomy. eLlife 10, e64325 (2021).
Miles-Jay, A. et al. Longitudinal genomic surveillance of carriage and transmission of Clostridioides difficile in an intensive care unit. Nat. Med. 29, 2526–2534 (2023).
Newcomer, E. P. et al. Genomic surveillance of Clostridioides difficile transmission and virulence in a healthcare setting. mBio 15, e0330023 (2024).
Coia, C. W., Banks, A. L., Cottom, L. & Fitzpatrick, F. The need for European surveillance of CDI. Adv. Exp. Med. Biol. 1435, 13–31 (2024).
UK Health Security Agency. Annual Epidemiological Commentary: Gram-Negative, MRSA, MSSA Bacteraemia and C. difficile Infections, Up to and Including Financial Year 2022 to 2023 (UKHSA, 2024).
Australian Commission on Safety and Quality in Health Care. Monitoring Clostridioides difficile Infection (CDI) in Australia. Clostridioides difficile Infection — 2020 and 2021 Data Snapshot (ACSQHC, 2023).
Choi, K. B. et al. Trends in Clostridioides difficile infection rates in Canadian hospitals during the coronavirus disease 2019 (COVID-19) pandemic. Infect. Control. Hosp. Epidemiol. 44, 1180–1183 (2023).
Karampatakis, T. et al. Implication of COVID-19 pandemic on the incidence of Clostridioides difficile infection in a Greek tertiary hospital. J. Med. Microbiol. https://doi.org/10.1099/jmm.0.001689 (2023).
Maldonado-Barrueco, A. et al. Increase of healthcare-onset Clostridioides difficile infection in adult population since SARS-CoV-2 pandemic: a retrospective cohort study in a tertiary care hospital from 2019 to 2022. Anaerobe 86, 102836 (2024).
Vehreschild, M. et al. Trends in the epidemiology of Clostridioides difficile infection in Germany. Infection 51, 1695–1702 (2023).
Tossens, B. et al. Impact of the COVID-19 pandemic on Clostridioides difficile infection in a tertiary healthcare institution in Belgium. Acta Clin. Belg. 78, 459–466 (2023).
Merchante, N. et al. Impact of COVID19 pandemic on the incidence of health-care associated Clostridioides difficile infection. Anaerobe 75, 102579 (2022).
European Centre for Disease Prevention and Control. Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals — 2002–2023 (ECDC, 2024).
Feuerstadt, P., Theriault, N. & Tillotson, G. The burden of CDI in the United States: a multifactorial challenge. BMC Infect. Dis. 23, 132 (2023).
Acknowledgements
C.H.C., C.N., I.B.M., J.F., K.D. and M.H.W. acknowledge the support of the National Institute for Health and Care Research (NIHR) Leeds Biomedical Research Centre (grant no. NIHR203331). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, the UK Health Security Agency (UKHSA) or the Department of Health and Social Care. M.H.W., K.D. and J.F acknowledge the support of the NIHR Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance (grant no. NIHR207397), a partnership between the UKHSA and the University of Oxford. M.H.W. and V.V. acknowledge the support of the NIHR Oxford Biomedical Research Centre and the NIHR Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance (grant no. NIHR200915), a partnership between the UKHSA and the University of Oxford. K.D. acknowledges the support of the NIHR HealthTech Research Centre in Accelerated Surgical Care.
Author information
Authors and Affiliations
Contributions
C.H.C. led the review and edit process of this article. The other authors contributed equally to all other aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
In the past 3 years, C.H.C. has received research funding from Debiopharm International and the MRC. I.B.M. has received research funding from GSK, Seres Therapeutics and the European Tissue Symposium. A.M.B. has received research funding from Seres Therapeutics. J.F. has received honorarium from Tillotts Pharma UK and grants from ACT-IVD, AJ Biosciences, Crestone, ESCMID, the MRC, the National Institute for Health and Care Research (NIHR), Pfizer and the UK Health Security Agency (UKHSA). K.D. has received honorarium from BD, Tillotts Pharma UK and Tillotts Pharma Europe, and grants from AJ Biosciences, ACT-IVD, AJ Biosciences, the MRC, the NIHR and Pfizer. M.H.W. has received consulting fees from AstraZeneca, Debiopharm, Ferring Pharmaceuticals, GSK, Nestlé, Paion, Pfizer, Phico Therapeutics, Qpex Biopharma, Seres Therapeutics, Summit, The European Tissue Symposium, Tillotts and Vedanta; lecture fees from GSK, Pfizer, Seres Therapeutics and Tillotts; and grant support from Debiopharm, GSK, Pfizer, Seres Therapeutics, Summit, the European Tissue Symposium and Tillotts. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Dena Lyras, who co-reviewed with Steven Mileto; Joseph Zackular; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chilton, C.H., Viprey, V., Normington, C. et al. Clostridioides difficile pathogenesis and control. Nat Rev Microbiol (2025). https://doi.org/10.1038/s41579-025-01242-2
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41579-025-01242-2