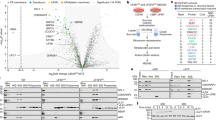
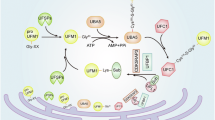
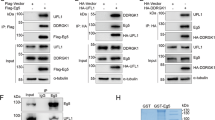
Abstract
UFMylation is a ubiquitin-like post-translational modification that has a central role in ribosome-associated quality control at the endoplasmic reticulum (ER-RQC). Through a dedicated enzymatic cascade, UFM1 is conjugated to select substrates, notably the 60S ribosomal subunit protein RPL26, to maintain endoplasmic reticulum and ribosomal integrity under cellular stress. This Review focuses on the structural and mechanistic basis of UFMylation in ER-RQC and its contribution to proteostasis. Although recent studies have identified a growing number of putative UFM1-modified proteins across diverse cellular pathways, the physiological importance of many of these substrates remains unclear. We highlight both the emerging functional breadth of UFMylation and the need for caution in interpreting substrate relevance. UFMylation is increasingly linked to disease, including neurodevelopmental disorders and cancer, underscoring its biological importance. Together, these findings position UFMylation as a key regulatory system connecting endoplasmic reticulum function to broader stress responses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006).
Cappadocia, L. & Lima, C. D. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 118, 889–918 (2018).
Gerakis, Y., Quintero, M., Li, H. & Hetz, C. The UFMylation system in proteostasis and beyond. Trends Cell Biol. 29, 974–986 (2019).
Millrine, D., Peter, J. J. & Kulathu, Y. A guide to UFMylation, an emerging posttranslational modification. FEBS J. 290, 5040–5056 (2023).
Komatsu, M., Inada, T. & Noda, N. N. The UFM1 system: working principles, cellular functions, and pathophysiology. Mol. Cell 84, 156–169 (2024).
Komatsu, M. et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 23, 1977–1986 (2004).
Tatsumi, K. et al. A novel type of E3 ligase for the Ufm1 conjugation system. J. Biol. Chem. 285, 5417–5427 (2010).
Peter, J. J. et al. A non-canonical scaffold-type E3 ligase complex mediates protein UFMylation. EMBO J. 41, e111015 (2022).
Yang, R. et al. CDK5RAP3, a UFL1 substrate adaptor, is crucial for liver development. Development 146, dev.169235 (2019).
Kang, S. H. et al. Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J. Biol. Chem. 282, 5256–5262 (2007).
Millrine, D. et al. Human UFSP1 is an active protease that regulates UFM1 maturation and UFMylation. Cell Rep. 40, 111168 (2022).
Scavone, F., Gumbin, S. C., Da Rosa, P. A. & Kopito, R. R. RPL26/uL24 UFMylation is essential for ribosome-associated quality control at the endoplasmic reticulum. Proc. Natl Acad. Sci. USA 120, e2220340120 (2023).
Walczak, C. P. et al. Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl Acad. Sci. USA 116, 1299–1308 (2019).
Wang, L. et al. SAYSD1 senses UFMylated ribosome to safeguard co-translational protein translocation at the endoplasmic reticulum. Cell Rep. 42, 112028 (2023).
Wang, L. et al. UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 30, 5–20 (2020).
Ishimura, R. et al. The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3. Nat. Commun. 13, 7857 (2022).
Stephani, M. et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. eLife 9, e58396 (2020).
Picchianti, L. et al. Shuffled ATG8 interacting motifs form an ancestral bridge between UFMylation and autophagy. EMBO J. 42, e112053 (2023).
Liang, J. R. et al. A genome-wide ER-phagy screen highlights key roles of mitochondrial metabolism and ER-resident UFMylation. Cell 180, 1160–1177.e20 (2020).
Li, B. et al. Ufmylation reconciles salt stress-induced unfolded protein responses via ER-phagy in Arabidopsis. Proc. Natl Acad. Sci. USA 120, e2208351120 (2023).
DaRosa, P. A. et al. UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER. Nature 627, 445–452 (2024).
Makhlouf, L. et al. The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons. Nature 627, 437–444 (2024).
Ishimura, R. et al. Mechanistic insights into the roles of the UFM1 E3 ligase complex in ufmylation and ribosome-associated protein quality control. Sci. Adv. 9, eadh3635 (2023).
Gong, Y. et al. PARP1 UFMylation ensures the stability of stalled replication forks. Proc. Natl Acad. Sci. USA 121, e2322520121 (2024).
Panichnantakul, P. et al. Protein UFMylation regulates early events during ribosomal DNA-damage response. Cell Rep. 43, 114738 (2024).
Yip, M. C. J. et al. Mechanism for recycling tRNAs on stalled ribosomes. Nat. Struct. Mol. Biol. 26, 343–349 (2019).
Qin, B. et al. STK38 promotes ATM activation by acting as a reader of histone H4 ufmylation. Sci. Adv. 6, eaax8214 (2020).
Yang, S., Moy, N. & Yang, R. The UFM1 conjugation system in mammalian development. Dev. Dyn. 252, 976–985 (2023).
Wang, R. N., Li, L., Zhou, J. & Ran, J. Multifaceted roles of UFMylation in health and disease. Acta Pharmacol. Sin. https://doi.org/10.1038/s41401-024-01456-9 (2025).
Colin, E. et al. Biallelic variants in UBA5 reveal that disruption of the UFM1 cascade can result in early-onset encephalopathy. Am. J. Hum. Genet. 99, 695–703 (2016).
Muona, M. et al. Biallelic variants in UBA5 link dysfunctional UFM1 ubiquitin-like modifier pathway to severe infantile-onset encephalopathy. Am. J. Hum. Genet. 99, 683–694 (2016).
Parra Bravo, C. et al. Human iPSC 4R tauopathy model uncovers modifiers of tau propagation. Cell 187, 2446–2464.e22 (2024).
Wang, L. et al. Mono-UFMylation promotes misfolding-associated secretion of α-synuclein. Sci. Adv. 10, eadk2542 (2024).
Sasakawa, H. et al. Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1. Biochem. Biophys. Res. Commun. 343, 21–26 (2006).
Bacik, J. P., Walker, J. R., Ali, M., Schimmer, A. D. & Dhe-Paganon, S. Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: mechanistic insights into a minimalistic E1 enzyme. J. Biol. Chem. 285, 20273–20280 (2010).
Oweis, W. et al. Trans-binding mechanism of ubiquitin-like protein activation revealed by a UBA5-UFM1 complex. Cell Rep. 16, 3113–3120 (2016).
Soudah, N. et al. An N-terminal extension to UBA5 adenylation domain boosts UFM1 activation: isoform-specific differences in ubiquitin-like protein activation. J. Mol. Biol. 431, 463–478 (2019).
Habisov, S. et al. Structural and functional analysis of a novel interaction motif within UFM1-activating enzyme 5 (UBA5) required for binding to ubiquitin-like proteins and ufmylation. J. Biol. Chem. 291, 9025–9041 (2016).
Kumar, M. et al. Structural basis for UFM1 transfer from UBA5 to UFC1. Nat. Commun. 12, 5708 (2021).
Wesch, N., Lohr, F., Rogova, N., Dotsch, V. & Rogov, V. V. A concerted action of UBA5 C-terminal unstructured regions is important for transfer of activated UFM1 to UFC1. Int. J. Mol. Sci. 22, 7390 (2021).
Taherbhoy, A. M. et al. Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol. Cell 44, 451–461 (2011).
Noda, N. N. et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 44, 462–475 (2011).
Lemaire, K. et al. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS ONE 6, e18517 (2011).
Zhang, Y., Zhang, M., Wu, J., Lei, G. & Li, H. Transcriptional regulation of the Ufm1 conjugation system in response to disturbance of the endoplasmic reticulum homeostasis and inhibition of vesicle trafficking. PLoS ONE 7, e48587 (2012).
Banerjee, S. et al. Structural study of UFL1-UFC1 interaction uncovers the role of UFL1 N-terminal helix in ufmylation. EMBO Rep. 24, e56920 (2023).
Liang, Q. et al. Human UFSP1 translated from an upstream near-cognate initiation codon functions as an active UFM1-specific protease. J. Biol. Chem. 298, 102016 (2022).
Ha, B. H. et al. Structure of ubiquitin-fold modifier 1-specific protease UfSP2. J. Biol. Chem. 286, 10248–10257 (2011).
Kim, K. H., Ha, B. H. & Kim, E. E. Structural basis for Ufm1 recognition by UfSP. FEBS Lett. 592, 263–273 (2018).
Penchev, I. et al. UFMylation orchestrates spatiotemporal coordination of RQC at the ER. Sci. Adv. 11, eadv0435 (2025).
Chen, C., Itakura, E., Weber, K. P., Hegde, R. S. & de Bono, M. An ER complex of ODR-4 and ODR-8/Ufm1 specific protease 2 promotes GPCR maturation by a Ufm1-independent mechanism. PLoS Genet. 10, e1004082 (2014).
Akinniyi, O. T. & Reese, J. C. DEF1: much more than an RNA polymerase degradation factor. DNA Repair 107, 103202 (2021).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
Martin, P. B. et al. NEMF mutations that impair ribosome-associated quality control are associated with neuromuscular disease. Nat. Commun. 11, 4625 (2020).
Inada, T. & Beckmann, R. Mechanisms of translation-coupled quality control. J. Mol. Biol. 436, 168496 (2024).
Joazeiro, C. A. P. Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol. Cell Biol. 20, 368–383 (2019).
Inada, T. Quality controls induced by aberrant translation. Nucleic Acids Res. 48, 1084–1096 (2020).
Brandman, O. & Hegde, R. S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 23, 7–15 (2016).
Inada, T. The ribosome as a platform for mRNA and nascent polypeptide quality control. Trends Biochem. Sci. 42, 5–15 (2017).
Sitron, C. S. & Brandman, O. Detection and degradation of stalled nascent chains via ribosome-associated quality control. Annu. Rev. Biochem. 89, 417–442 (2020).
Mochida, K. & Nakatogawa, H. ER-phagy: selective autophagy of the endoplasmic reticulum. EMBO Rep. 23, e55192 (2022).
Reggiori, F. & Molinari, M. ER-phagy: mechanisms, regulation, and diseases connected to the lysosomal clearance of the endoplasmic reticulum. Physiol. Rev. 102, 1393–1448 (2022).
Chino, H. & Mizushima, N. ER-phagy: quality and quantity control of the endoplasmic reticulum by autophagy. Cold Spring Harb. Perspect. Biol. 15, a041256 (2023).
Li, J. et al. Ufm1-specific ligase Ufl1 regulates endoplasmic reticulum homeostasis and protects against heart failure. Circ. Heart Fail. 11, e004917 (2018).
Zhou, Y. et al. Ufl1 deficiency causes kidney atrophy associated with disruption of endoplasmic reticulum homeostasis. J. Genet. Genom. 48, 403–410 (2021).
Miller, C. et al. RCAD/BiP pathway is necessary for the proper synthesis of digestive enzymes and secretory function of the exocrine pancreas. Am. J. Physiol. Gastrointest. Liver Physiol. 312, G314–G326 (2017).
Wang, Z. et al. MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 47, 4124–4135 (2019).
Lee, L. et al. UFMylation of MRE11 is essential for telomere length maintenance and hematopoietic stem cell survival. Sci. Adv. 7, eabc7371 (2021).
Tian, T. et al. UFL1 triggers replication fork degradation by MRE11 in BRCA1/2-deficient cells. Nat. Chem. Biol. 20, 1650–1661 (2024).
Qin, B. et al. UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 10, 1242 (2019).
Tan, Q. & Xu, X. PTIP UFMylation promotes replication fork degradation in BRCA1-deficient cells. J. Biol. Chem. 300, 107312 (2024).
Liu, J. et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat. Cell Biol. 22, 1056–1063 (2020).
Snider, D. L., Park, M., Murphy, K. A., Beachboard, D. C. & Horner, S. M. Signaling from the RNA sensor RIG-I is regulated by ufmylation. Proc. Natl Acad. Sci. USA 119, e2119531119 (2022).
Yiu, S. P. T. et al. An Epstein-Barr virus protein interaction map reveals NLRP3 inflammasome evasion via MAVS UFMylation. Mol. Cell 83, 2367–2386.e15 (2023).
Zhou, J. et al. Dysregulation of PD-L1 by UFMylation imparts tumor immune evasion and identified as a potential therapeutic target. Proc. Natl Acad. Sci. USA 120, e2215732120 (2023).
He, C. et al. UFL1 ablation in T cells suppresses PD-1 UFMylation to enhance anti-tumor immunity. Mol. Cell 84, 1120–1138.e8 (2024).
Mao, M. et al. Modification of PLAC8 by UFM1 affects tumorous proliferation and immune response by impacting PD-L1 levels in triple-negative breast cancer. J. Immunother. Cancer 10, e005668 (2022).
Zhu, J. et al. P4HB UFMylation regulates mitochondrial function and oxidative stress. Free Radic. Biol. Med. 188, 277–286 (2022).
Yang, J. et al. Metformin induces ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J. Exp. Clin. Cancer Res. 40, 206 (2021).
Turano, C., Coppari, S., Altieri, F. & Ferraro, A. Proteins of the PDI family: unpredicted non-ER locations and functions. J. Cell Physiol. 193, 154–163 (2002).
Cai, Y. et al. UFBP1, a key component of the Ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLoS Genet. 11, e1005643 (2015).
Tatsumi, K. et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2, 181 (2011).
Zhang, M. et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 22, 1922–1934 (2015).
Egunsola, A. T. et al. Loss of DDRGK1 modulates SOX9 ubiquitination in spondyloepimetaphyseal dysplasia. J. Clin. Invest. 127, 1475–1484 (2017).
Cai, Y. et al. Indispensable role of the ubiquitin-fold modifier 1-specific E3 ligase in maintaining intestinal homeostasis and controlling gut inflammation. Cell Discov. 5, 7 (2019).
Quintero, M. et al. Cdk5rap3 is essential for intestinal Paneth cell development and maintenance. Cell Death Dis. 12, 131 (2021).
Balce, D. R. et al. UFMylation inhibits the proinflammatory capacity of interferon-γ-activated macrophages. Proc. Natl Acad. Sci. USA 118, e2011763118 (2021).
Zhou, X. et al. UFMylation: a ubiquitin-like modification. Trends Biochem. Sci. 49, 52–67 (2024).
Nahorski, M. S. et al. Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development. Brain 141, 1934–1945 (2018).
Zhang, J. et al. Deficiency of murine UFM1-specific E3 ligase causes microcephaly and inflammation. Mol. Neurobiol. 59, 6363–6372 (2022).
Zhang, G. et al. UFSP2-related spondyloepimetaphyseal dysplasia: a confirmatory report. Eur. J. Med. Genet. 63, 104021 (2020).
Di Rocco, M. et al. Novel spondyloepimetaphyseal dysplasia due to UFSP2 gene mutation. Clin. Genet. 93, 671–674 (2018).
Watson, C. M. et al. Identification of a mutation in the ubiquitin-fold modifier 1-specific peptidase 2 gene, UFSP2, in an extended South African family with Beukes hip dysplasia. S. Afr. Med. J. 105, 558–563 (2015).
Zhou, J. et al. Genomic profiling of the UFMylation family genes identifies UFSP2 as a potential tumour suppressor in colon cancer. Clin. Transl. Med. 11, e642 (2021).
Fang, B., Li, Z., Qiu, Y., Cho, N. & Yoo, H. M. Inhibition of UBA5 expression and induction of autophagy in breast cancer cells by usenamine A. Biomolecules 11, 1348 (2021).
Chen, F. et al. Loss of Ufl1/Ufbp1 in hepatocytes promotes liver pathological damage and carcinogenesis through activating mTOR signaling. J. Exp. Clin. Cancer Res. 42, 110 (2023).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Grau-Bove, X., Sebe-Pedros, A. & Ruiz-Trillo, I. The eukaryotic ancestor had a complex ubiquitin signaling system of archaeal origin. Mol. Biol. Evol. 32, 726–739 (2015).
Kwon, Y. T. & Ciechanover, A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 42, 873–886 (2017).
Dikic, I. & Schulman, B. A. An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 24, 273–287 (2023).
Vertegaal, A. C. O. Signalling mechanisms and cellular functions of SUMO. Nat. Rev. Mol. Cell Biol. 23, 715–731 (2022).
Harper, J. W. & Schulman, B. A. Cullin-RING ubiquitin ligase regulatory circuits: a quarter century beyond the F-box hypothesis. Annu. Rev. Biochem. 90, 403–429 (2021).
Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 63, 1–10 (2020).
Zhang, X. & Chen, X. L. The emerging roles of ubiquitin-like protein Urm1 in eukaryotes. Cell Signal. 81, 109946 (2021).
Hermann, M. & Bogunovic, D. ISG15: in sickness and in health. Trends Immunol. 38, 79–93 (2017).
Ye, Y. & Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 (2009).
Zheng, N. & Shabek, N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017).
Furukawa, K., Mizushima, N., Noda, T. & Ohsumi, Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 275, 7462–7465 (2000).
Trempe, J. F. Reading the ubiquitin postal code. Curr. Opin. Struct. Biol. 21, 792–801 (2011).
Yau, T. Y., Sander, W., Eidson, C. & Courey, A. J. SUMO interacting motifs: structure and function. Cells 10, 2825 (2021).
Noda, N. N., Ohsumi, Y. & Inagaki, F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385 (2010).
Birgisdottir, A. B., Lamark, T. & Johansen, T. The LIR motif — crucial for selective autophagy. J. Cell Sci. 126, 3237–3247 (2013).
Kolla, S., Ye, M., Mark, K. G. & Rape, M. Assembly and function of branched ubiquitin chains. Trends Biochem. Sci. 47, 759–771 (2022).
Vertegaal, A. C. SUMO chains: polymeric signals. Biochem. Soc. Trans. 38, 46–49 (2010).
Enchev, R. I., Schulman, B. A. & Peter, M. Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 (2015).
Yoo, H. M. et al. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 56, 261–274 (2014).
Acknowledgements
This work was supported by JSPS KAKENHI grant numbers 23K20044 and 24H00060 (to M.K.), JP25H01320 (to N.N.N.) and JP25H00007 (to T.I.). The authors thank their laboratory colleagues and Y. Dagdas for the insightful discussions. This work is dedicated to the memory of K. Tanaka, who passed away on 23 July 2024, in recognition of his profound contributions to the field.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Yafei Cai and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Ferroptosis
-
A regulated, iron-dependent form of cell death driven by uncontrolled lipid peroxidation, arising from failure of the glutathione-dependent antioxidant system, and genetically and biochemically distinct from apoptosis and other programmed death pathways.
- Life history strategies
-
Patterns of growth, reproduction and survival that characterize how an organism allocates resources across its lifespan.
- Listerin
-
(LTN1). An E3 ubiquitin ligase in the ribosome-associated quality control (RQC) pathway that ubiquitinates nascent polypeptides on stalled ribosomes to target them for degradation.
- Nuclear export mediator factor
-
(NEMF). A core component of the RQC machinery that recognizes stalled ribosomes and recruits downstream RQC factors.
- PINK1–Parkin pathway
-
A mitochondrial quality control pathway in which PINK1 accumulates on damaged mitochondria and recruits or activates the E3 ligase Parkin, triggering mitophagy — the selective autophagic removal of damaged mitochondria.
- RQC factors
-
Components of the ribosome-associated quality control pathway that detect stalled or collided ribosomes and mediate the ubiquitination, extraction or degradation of incomplete nascent chains after the dissociation of ribosomes.
- SEC61 translocon
-
A protein-conducting channel in the endoplasmic reticulum membrane that mediates the co-translational translocation of nascent polypeptides into or across the ER membrane.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Komatsu, M., Noda, N.N. & Inada, T. The mechanistic basis and cellular functions of UFMylation. Nat Rev Mol Cell Biol (2026). https://doi.org/10.1038/s41580-025-00944-y
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41580-025-00944-y