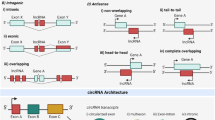
Abstract
Tens, if not hundreds, of thousands of long non-coding RNAs (lncRNAs) are transcribed from mammalian genomes, especially in the brain, wherein most exhibit region-specific and/or cell-specific expression patterns. Many lncRNAs are nuclear-localized and appear to be the products of developmental enhancers, whereas others are found in the cytoplasm, including at the synapse. Here, we describe the lncRNAs that have been shown to have roles in various aspects of brain development, synaptic function, learning, behaviour and brain disorders. Our emerging understanding indicates that lncRNAs direct many, if not most, of the regulatory transactions that give rise to the structure of the brain and modulate its functions, probably through their guidance of relatively generic effector proteins. Although they hold promise as targets for therapeutic interventions, a concerted effort will be required to characterize the structures, functions, spatial distribution and interacting partners of the lncRNAs expressed in the brain, most of which have not been studied. We suggest that the lncRNAs transcribed from genomic regions associated with human neurological traits and disorders be prioritized for analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Mattick, J. S. The central role of RNA in human development and cognition. FEBS Lett. 585, 1600–1616 (2011).
Barry, G. & Mattick, J. S. The role of regulatory RNA in cognitive evolution. Trends Cogn. Sci. 16, 497–503 (2012).
Ramos, A. D., Attenello, F. J. & Lim, D. A. Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci. Lett. 625, 70–79 (2016).
Ng, S.-Y., Lin, L., Soh, B. S. & Stanton, L. W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 29, 461–468 (2013).
Statello, L., Guo, C.-J., Chen, L.-L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021). This review outlines how lncRNAs regulate gene expression through their localization and interactions, highlighting their importance in cell differentiation and disorders.
Yang, S., Lim, K.-H., Kim, S.-H. & Joo, J.-Y. Molecular landscape of long noncoding RNAs in brain disorders. Mol. Psych. 26, 1060–1074 (2021).
Mattick, J. S. et al. Long noncoding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430–447 (2023). This paper presents the current consensus on the definition and functions of lncRNAs.
Ravasi, T. et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 16, 11–19 (2006).
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F. & Mattick, J. S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA 105, 716–712 (2008). This study shows that many lncRNAs are specifically expressed in distinct brain regions.
Liau, W.-S. et al. Fear extinction is regulated by the activity of long noncoding RNAs at the synapse. Nat. Commun. 14, 7616 (2023). This paper shows that many long lncRNAs are localized at the synapse and establishes their functional relevance at the subcellular level.
Tsagakis, I., Douka, K., Birds, I. & Aspden, J. L. Long non-coding RNAs in development and disease: conservation to mechanisms. J. Pathol. 250, 480–495 (2020).
Korneev, S. & O’Shea, M. Natural antisense RNAs in the nervous system. Rev. Neurosci. 16, 213–222 (2005).
Smalheiser, N. R. et al. Natural antisense transcripts are co-expressed with sense mRNAs in synaptoneurosomes of adult mouse forebrain. Neurosci. Res. 62, 236–239 (2008).
Rybak-Wolf, A. et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885 (2015).
You, X. et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610 (2015).
Gokool, A., Anwar, F. & Voineagu, I. The landscape of circular RNA expression in the human brain. Biol. Psych. 87, 294–304 (2020).
Walsh, K., Gokool, A., Alinejad-Rokny, H. & Voineagu, I. NeuroCirc: an integrative resource of circular RNA expression in the human brain. Bioinformatics 37, 3664–3666 (2021).
Zajaczkowski, E. L. et al. Localised Cdr1as activity is required for fear extinction memory. Neurobiol. Learn. Mem. 203, 107777 (2023).
Milligan, M. J. & Lipovich, L. Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front. Genet. 5, 476 (2015).
Cheetham, S. W., Faulkner, G. J. & Dinger, M. E. Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat. Rev. Genet. 21, 191–201 (2020).
Gonzàlez-Porta, M., Frankish, A., Rung, J., Harrow, J. & Brazma, A. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 14, R70 (2013). This paper shows that in almost 20% of protein-coding genes, the major transcript does not code for a protein.
Mercer, T. R. et al. Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res. 39, 2393–2403 (2011). This paper shows that there is widespread independent expression of 5′-capped 3′ untranslated regions in specific cells, and that these lncRNAs have regulatory functions in cell differentiation.
Kocabas, A., Duarte, T., Kumar, S. & Hynes, M. A. Widespread differential expression of coding region and 3′UTR sequences in neurons and other tissues. Neuron 88, 1149–1156 (2015).
Mattick, J. S. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. BioEssays 25, 930–939 (2003).
Kapranov, P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488 (2007).
Dinger, M. E., Gascoigne, D. K. & Mattick, J. S. The evolution of RNAs with multiple functions. Biochimie 93, 2013–2018 (2011).
Wu, P. et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 19, 22 (2020).
Senís, E. et al. TUNAR lncRNA encodes a microprotein that regulates neural differentiation and neurite formation by modulating calcium dynamics. Front. Cell Dev. Biol. 9, 747667 (2021).
Mercer, T. R. et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 20, 1639–1650 (2010).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012).
Carninci, P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005).
Clark, M. B. et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 21, 885–898 (2012).
Tani, H. et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 22, 947–956 (2012).
Shi, K., Liu, T., Fu, H., Li, W. & Zheng, X. Genome-wide analysis of lncRNA stability in human. PLoS Comput. Biol. 17, e1008918 (2021).
Quinn, J. J. & Chang, H. Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62 (2016). This review provides a comprehensive understanding of lncRNA biogenesis, structure and function.
Frith, M. C. et al. Evolutionary turnover of mammalian transcription start sites. Genome Res. 16, 713–722 (2006).
Phan, M. H. Q. et al. Conservation of regulatory elements with highly diverged sequences across large evolutionary distances. Nat. Genet. 57, 1524–1534 (2025). This paper shows that lack of primary sequence conservation is not an indicator of lack of regulatory function.
Kutter, C. et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 8, e1002841 (2012).
Pheasant, M. & Mattick, J. S. Raising the estimate of functional human sequences. Genome Res. 17, 1245–1253 (2007).
Ponjavic, J., Ponting, C. P. & Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 17, 556–565 (2007).
Smith, M. A., Gesell, T., Stadler, P. F. & Mattick, J. S. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res. 41, 8220–8236 (2013).
Nitsche, A., Rose, D., Fasold, M., Reiche, K. & Stadler, P. F. Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved. RNA 21, 801–812 (2015).
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H. & Bartel, D. P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550 (2011). This paper shows that lncRNAs with low primary sequence conservation (relative to proteins) can retain orthologous regulatory functions.
Quinn, J. J. et al. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev. 30, 191–207 (2016).
Degani, N., Lubelsky, Y., Perry, R. B.-T., Ainbinder, E. & Ulitsky, I. Highly conserved and cis-acting lncRNAs produced from paralogous regions in the center of HOXA and HOXB clusters in the endoderm lineage. PLoS Genet. 17, e1009681 (2021).
Santos-Rodriguez, G., Voineagu, I. & Weatheritt, R. J. Evolutionary dynamics of circular RNAs in primates. eLife 10, e69148 (2021).
Ross, C. J. & Ulitsky, I. Discovering functional motifs in long noncoding RNAs. WIREs RNA 13, e1708 (2022).
Li, S. et al. Gain of bipolar disorder-related lncRNA AP1AR-DT in mice induces depressive and anxiety-like behaviors by reducing Negr1-mediated excitatory synaptic transmission. BMC Med. 22, 543 (2024).
Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011).
Rinn, J. L. & Chang, H. Y. Long noncoding RNAs: molecular modalities to organismal functions. Annu. Rev. Biochem. 89, 283–308 (2020). This review describes how structure–function relationships in lncRNAs are linked to organism-level outcomes.
Leontis, N. B., Lescoute, A. & Westhof, E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 16, 279–287 (2006).
Westhof, E. & Fritsch, V. The endless subtleties of RNA-protein complexes. Structure 19, 902–903 (2011).
Morrissy, A. S., Griffith, M. & Marra, M. A. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 21, 1203–1212 (2011).
Werner, A., Kanhere, A., Wahlestedt, C. & Mattick, J. S. Natural antisense transcripts as versatile regulators of gene expression. Nat. Rev. Genet. 25, 730–744 (2024).
Ang, C. E., Trevino, A. E. & Chang, H. Y. Diverse lncRNA mechanisms in brain development and disease. Curr. Opin. Genet. Dev. 65, 42–46 (2020).
Somasundaram, K., Gupta, B., Jain, N. & Jana, S. LncRNAs divide and rule: the master regulators of phase separation. Front. Genet. 13, 930792 (2022).
Mattick, J. S. Enhancers are genes that express organizational RNAs. Front. RNA Res. 1, 1194526 (2023). This article summarizes the evidence that enhancer loci are genes producing long noncoding RNAs that regulate cell fate and development.
Cabili, M. N. et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16, 20 (2015). This study demonstrates that most long noncoding RNAs are localized in punctate, probably phase-separated, domains in cells.
Bridges, M. C., Daulagala, A. C. & Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 220, e202009045 (2021).
Carlevaro-Fita, J. & Johnson, R. Global positioning system: understanding long noncoding RNAs through subcellular localization. Mol. Cell 73, 869–883 (2019).
Liau, W.-S., Samaddar, S., Banerjee, S. & Bredy, T. W. On the functional relevance of spatiotemporally-specific patterns of experience-dependent long noncoding RNA expression in the brain. RNA Biol. 18, 1025–1036 (2021).
Nakagawa, S. & Hirose, T. Paraspeckle nuclear bodies — useful uselessness? Cell. Mol. Life Sci. 69, 3027–3036 (2012).
Yamazaki, T. et al. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053 (2018).
Ingram, H. B. & Fox, A. H. Unveiling the intricacies of paraspeckle formation and function. Curr. Opin. Cell Biol. 90, 102399 (2024).
Barry, G. et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci. Rep. 7, 40127 (2017).
Butler, A. A., Johnston, D. R., Kaur, S. & Lubin, F. D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 12, eaaw9277 (2019).
Kukharsky, M. S. et al. Long non-coding RNA Neat1 regulates adaptive behavioural response to stress in mice. Transl. Psych. 10, 171 (2020).
Zhang, B. et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123 (2012).
Balaji, A. et al. MALAT1 regulates mRNA processing through sequence dependent RNA-RNA and RNA-protein interactions. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaf784 (2025).
Kuo, P.-H., Doudeva, L. G., Wang, Y.-T., Shen, C.-K. J. & Yuan, H. S. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 37, 1799–1808 (2009).
Imre, L. et al. Nucleosome destabilization by polyamines. Arch. Biochem. Biophys. 722, 109184 (2022).
Gupta, V. K. et al. Restoring polyamines protects from age-induced memory impairment in an autophagy-dependent manner. Nat. Neurosci. 16, 1453–1460 (2013).
Chen, L. et al. Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J. Cell. Mol. Med. 20, 2102–2110 (2016).
Jiang, T. et al. The lncRNA MALAT1/miR-30/Spastin axis regulates hippocampal neurite outgrowth. Front. Cell. Neurosci. 14, 555747 (2020).
Kryger, R., Fan, L., Wilce, P. A. & Jaquet, V. MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol 46, 629–634 (2012).
Zhang, X., Tang, X., Liu, K., Hamblin, M. H. & Yin, K.-J. Long noncoding RNA Malat1 regulates cerebrovascular pathologies in ischemic stroke. J. Neurosci. 37, 1797–1806 (2017).
Xiao, W. et al. The lncRNA Malat1 is trafficked to the cytoplasm as a localized mRNA encoding a small peptide in neurons. Genes Dev. 38, 294–307 (2024).
Madugalle, S. U. et al. Synapse-enriched m6A-modified Malat1 interacts with the novel m6A reader, DPYSL2, and is required for fear-extinction memory. J. Neurosci. 43, 7084–7100 (2023). This study shows that synaptic lncRNAs and RNA modifications have a role in memory.
Mercer, T. R. et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11, 14 (2010).
Sone, M. et al. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 120, 2498–2506 (2007).
Rapicavoli, N. A., Poth, E. M. & Blackshaw, S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 10, 49 (2010).
Ishizuka, A., Hasegawa, Y., Ishida, K., Yanaka, K. & Nakagawa, S. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Genes Cell 19, 704–721 (2014).
Barry, G. et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psych. 19, 486–494 (2014).
Ishii, N. et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 51, 1087–1099 (2006).
Spadaro, P. A. et al. Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol. Psych. 78, 848–859 (2015).
Ip, J. Y. et al. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci. Rep. 6, 27204 (2016).
Michelhaugh, S. K. et al. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J. Neurochem. 116, 459–466 (2011).
Rao, S.-Q., Hu, H.-L., Ye, N., Shen, Y. & Xu, Q. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr. Res. 166, 125–130 (2015).
Teng, P. et al. The human lncRNA GOMAFU suppresses neuronal interferon response pathways affected in neuropsychiatric diseases. Brain Behav. Immun. 112, 175–187 (2023).
Grzejda, D. et al. The long noncoding RNA mimi scaffolds neuronal granules to maintain nervous system maturity. Sci. Adv. 8, eabo5578 (2022).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011).
Liu, S. J. et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 17, 67 (2016).
Ma, Q. & Chang, H. Y. Single-cell profiling of lncRNAs in the developing human brain. Genome Biol. 17, 68 (2016).
Deveson, I. W., Hardwick, S. A., Mercer, T. R. & Mattick, J. S. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 33, 464–478 (2017). This paper explains why lncRNAs are undersampled in many RNA-sequencing experiments.
Goff, L. A. et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA 112, 6855–6862 (2015).
Kadakkuzha, B. M. et al. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front. Cell. Neurosci. 9, 63 (2015).
Bocchi, V. D. et al. The coding and long noncoding single-cell atlas of the developing human fetal striatum. Science 372, eabf5759 (2021).
Navandar, M., Vennin, C., Lutz, B. & Gerber, S. Long non-coding RNAs expression and regulation across different brain regions in primates. Sci. Data 11, 545 (2024).
De Santa, F. et al. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol. 8, e1000384 (2010).
Orom, U. A. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58 (2010).
Li, W., Notani, D. & Rosenfeld, M. G. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 17, 207–223 (2016).
Sartorelli, V. & Lauberth, S. M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 27, 521–528 (2020).
Furlong, E. E. M. & Levine, M. Developmental enhancers and chromosome topology. Science 361, 1341–1345 (2018).
Hahn, S. Phase separation, protein disorder, and enhancer function. Cell 175, 1723–1725 (2018).
Popay, T. M. & Dixon, J. R. Coming full circle: on the origin and evolution of the looping model for enhancer–promoter communication. J. Biol. Chem. 298, 102117 (2022).
Schoenfelder, S. & Fraser, P. Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
Ramirez, M. et al. Identification and characterization of transcribed enhancers during cerebellar development through enhancer RNA analysis. BMC Genom. 24, 351 (2023).
Capauto, D. et al. Characterization of enhancer activity in early human neurodevelopment using massively parallel reporter assay (MPRA) and forebrain organoids. Sci. Rep. 14, 3936 (2024).
Yoshihara, M. et al. Transcriptional enhancers in human neuronal differentiation provide clues to neuronal disorders. EMBO Rep. 26, 1212–1237 (2025).
Ulitsky, I. & Bartel, D. P. lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46 (2013).
Necsulea, A. et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640 (2014).
Reilly, S. K. et al. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347, 1155–1159 (2015).
Zimmer-Bensch, G. Emerging roles of long non-coding RNAs as drivers of brain evolution. Cells 8, 1399 (2019).
Mangan, R. J. et al. Adaptive sequence divergence forged new neurodevelopmental enhancers in humans. Cell 185, 4587–4603 (2022).
Prabhakar, S. et al. Human-specific gain of function in a developmental enhancer. Science 321, 1346–1350 (2008).
Gasperini, M., Tome, J. M. & Shendure, J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 21, 292–310 (2020).
Dong, X. et al. Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat. Neurosci. 21, 1482–1492 (2018).
Shima, Y. et al. A mammalian enhancer trap resource for discovering and manipulating neuronal cell types. eLife 5, e13503 (2016).
Kim, T.-K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Wei, W. et al. ADRAM is an experience-dependent long noncoding RNA that drives fear extinction through a direct interaction with the chaperone protein 14-3-3. Cell Rep. 38, 110546 (2022).
Griffith, E. C., West, A. E. & Greenberg, M. E. Neuronal enhancers fine-tune adaptive circuit plasticity. Neuron 112, 3043–3057 (2024).
Ptashne, M. How eukaryotic transcriptional activators work. Nature 335, 683–689 (1988).
Orom, U. A. & Shiekhattar, R. Noncoding RNAs and enhancers: complications of a long-distance relationship. Trends Genet. 27, 433–439 (2011).
Mercer, T. R. & Mattick, J. S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307 (2013).
Soibam, B. Super-lncRNAs: identification of lncRNAs that target super-enhancers via RNA:DNA:DNA triplex formation. RNA 23, 1729–1742 (2017).
Li, Y., Syed, J. & Sugiyama, H. RNA-DNA triplex formation by long noncoding RNAs. Cell Chem. Biol. 23, 1325–1333 (2016).
Isoda, T. et al. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell 171, 103–119 (2017).
Cajigas, I. et al. Sox2-Evf2 lncRNA mechanisms of chromosome topological control in developing forebrain. Development 148, dev197202 (2021).
Cetin, N. S. et al. Isolation and genome-wide characterization of cellular DNA:RNA triplex structures. Nucleic Acids Res. 47, 2306–2321 (2019).
Arnold, P. R., Wells, A. D. & Li, X. C. Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate. Front. Cell Dev. Biol. 7, 377 (2020).
Soibam, B. & Zhamangaraeva, A. LncRNA:DNA triplex-forming sites are positioned at specific areas of genome organization and are predictors for topologically associated domains. BMC Genom. 22, 397 (2021).
Guttman, M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011).
Flynn, R. A. & Chang, H. Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14, 752–761 (2014). This paper provides an excellent review of the accumulating evidence that lncRNAs regulate cell differentiation.
Deveson, I. W. et al. Universal alternative splicing of noncoding exons. Cell Syst. 6, 245–255 (2018).
Maass, P. G. et al. A misplaced lncRNA causes brachydactyly in humans. J. Clin. Invest. 122, 3990–4002 (2012).
Melo, C. A. et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell 49, 524–535 (2013).
Li, W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013).
Xiang, J. F. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 24, 513–531 (2014).
Paralkar, V. R. et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood 123, 1927–1937 (2014).
Yin, Y. et al. Opposing roles for the lncRNA Haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16, 504–516 (2015).
Yang, Y. et al. Enhancer RNA-driven looping enhances the transcription of the long noncoding RNA DHRS4-AS1, a controller of the DHRS4 gene cluster. Sci. Rep. 6, 20961 (2016).
Fatima, R., Choudhury, S. R., T R, D., Bhaduri, U. & Rao, M. R. S. A novel enhancer RNA, Hmrhl, positively regulates its host gene, phkb, in chronic myelogenous leukemia. Noncoding RNA Res. 4, 96–108 (2019).
Lewandowski, J. P. et al. The Firre locus produces a trans-acting RNA molecule that functions in hematopoiesis. Nat. Commun. 10, 5137 (2019).
Setten, R. L., Chomchan, P., Epps, E. W., Burnett, J. C. & Rossi, J. J. CRED9: a differentially expressed elncRNA regulates expression of transcription factor CEBPA. RNA 27, 891–906 (2021).
Alvarez-Dominguez, J. R., Knoll, M., Gromatzky, A. A. & Lodish, H. F. The super-enhancer-derived alncRNA-EC7/Bloodlinc potentiates red blood cell development in trans. Cell Rep. 19, 2503–2514 (2017).
Shii, L., Song, L., Maurer, K., Zhang, Z. & Sullivan, K. E. SERPINB2 is regulated by dynamic interactions with pause-release proteins and enhancer RNAs. Mol. Immunol. 88, 20–31 (2017).
Gil, N. & Ulitsky, I. Production of spliced long noncoding RNAs specifies regions with increased enhancer activity. Cell Syst. 7, 537–547 (2018).
Tan, J. Y., Biasini, A., Young, R. S. & Marques, A. C. Splicing of enhancer-associated lincRNAs contributes to enhancer activity. Life Sci. Alliance 3, e202000663 (2020).
Tan, J. Y. & Marques, A. C. The activity of human enhancers is modulated by the splicing of their associated lncRNAs. PLoS Comput. Biol. 18, e1009722 (2022). This is one of several papers indicating that lncRNAs mediate enhancer activity.
Lee, J.-H. et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell 81, 3368–3385 (2021).
Pang, K. C., Frith, M. C. & Mattick, J. S. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 22, 1–5 (2006).
Hezroni, H. et al. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 11, 1110–1122 (2015).
Unfried, J. P. & Ulitsky, I. Substoichiometric action of long noncoding RNAs. Nat. Cell Biol. 24, 608–615 (2022).
Rani, N. et al. A primate lncRNA mediates Notch signaling during neuronal development by sequestering miRNA. Neuron 90, 1174–1188 (2016).
Bond, A. M. et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027 (2009).
Kohtz, J. et al. The Evf2 enhancer lncRNA links chromosomal interactions and interneuron diversity. FASEB J. 31, 128.1 (2017).
Feng, J. et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484 (2006).
Cajigas, I. et al. The Evf2 ultraconserved enhancer lncRNA functionally and spatially organizes megabase distant genes in the developing forebrain. Mol. Cell 71, 956–972 (2018). This study shows how an enhancer-derived lncRNA organizes chromatin topology to regulate distal genes involved in interneuron diversity and seizure susceptibility.
Berghoff, E. G. et al. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 140, 4407–4416 (2013).
Cajigas, I. et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development 142, 2641–2652 (2015).
Sierra, C. et al. The lncRNA Snhg11, a new candidate contributing to neurogenesis, plasticity, and memory deficits in Down syndrome. Mol. Psych. 29, 2117–2134 (2024).
Ang, C. E. et al. The novel lncRNA lnc-NR2F1 is pro-neurogenic and mutated in human neurodevelopmental disorders. eLife 8, e41770 (2019).
Wei, M. et al. Axon-enriched lincRNA ALAE is required for axon elongation via regulation of local mRNA translation. Cell Rep. 35, 109053 (2021).
Chen, H.-S. et al. Long noncoding RNA Gm2694 drives depressive-like behaviors in male mice by interacting with GRP78 to disrupt endoplasmic reticulum homeostasis. Sci. Adv. 8, eabn2496 (2022).
Wang, F. et al. The long noncoding RNA Synage regulates synapse stability and neuronal function in the cerebellum. Cell Death Diff. 28, 2634–2650 (2021).
Carrieri, C. et al. Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front. Cell. Neurosci. 9, 114 (2015).
Maag, J. L. V. et al. Dynamic expression of long noncoding RNAs and repeat elements in synaptic plasticity. Front. Neurosci. 9, 351 (2015).
Chen, B. J. et al. RNA sequencing reveals pronounced changes in the noncoding transcriptome of aging synaptosomes. Neurobiol. Aging 56, 67–77 (2017).
Cui, X. et al. Identification and characterization of long non-coding RNA Carip in modulating spatial learning and memory. Cell Rep. 38, 110398 (2022).
Grinman, E. et al. Activity-regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites. Sci. Adv. 7, eabf0605 (2021).
Raveendra, B. L. et al. Long noncoding RNA GM12371 acts as a transcriptional regulator of synapse function. Proc. Natl Acad. Sci. USA 115, E10197–E10205 (2018).
Li, D. et al. Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat. Commun. 9, 1726 (2018).
Kelly, D. et al. A functional screen uncovers circular RNAs regulating excitatory synaptogenesis in hippocampal neurons. Nat. Commun. 16, 3040 (2025).
Silenzi, V. et al. A tripartite circRNA/mRNA/miRNA interaction regulates glutamatergic signaling in the mouse brain. Cell Rep. 43, 114766 (2024).
Ramos, A. D. et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16, 439–447 (2015).
Saha, P. et al. Sex-specific role for the long noncoding RNA Pnky in mouse behavior. Nat. Commun. 15, 6901 (2024).
Issler, O. et al. Sex-specific role for the long non-coding RNA LINC00473 in depression. Neuron 106, 912–926 (2020).
Issler, O. et al. The long noncoding RNA FEDORA is a cell type- and sex-specific regulator of depression. Sci. Adv. 8, eabn9494 (2022).
Xu, H. et al. Role of long noncoding RNA Gas5 in cocaine action. Biol. Psych. 88, 758–766 (2020).
Zhao, H. et al. GAS5 which is regulated by Lhx8 promotes the recovery of learning and memory in rats with cholinergic nerve injury. Life Sci. 260, 118388 (2020).
Ma, M. et al. A novel pathway regulates social hierarchy via lncRNA AtLAS and postsynaptic synapsin IIb. Cell Res. 30, 105–118 (2020).
Riva, P., Ratti, A. & Venturin, M. The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr. Alzheimer Res. 13, 1219–1231 (2016).
Kallweit, L. et al. Chronic RNA G-quadruplex accumulation in aging and Alzheimer’s disease. eLife 14, RP105446 (2025).
Bhagat, R. et al. Long non-coding RNA SNHG8 drives stress granule formation in tauopathies. Mol. Psych. 28, 4889–4901 (2023).
Parikshak, N. N. et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427 (2016).
Sparber, P., Filatova, A., Khantemirova, M. & Skoblov, M. The role of long non-coding RNAs in the pathogenesis of hereditary diseases. BMC Med. Genom. 12, 42 (2019).
Chen, K.-W. & Chen, J.-A. Functional roles of long non-coding RNAs in motor neuron development and disease. J. Biomed. Sci. 27, 38 (2020).
Ganesh, V. S. et al. Neurodevelopmental disorder caused by deletion of CHASERR, a lncRNA gene. N. Engl. J. Med. 391, 1511–1518 (2024). This paper shows that a large deletion produces a developmental disorder, consistent with the role of lncRNAs in regulating development and cell differentiation, whereas it is probable that smaller variations are responsible for quantitative trait variation.
Li, P. P. et al. ATXN2-AS, a gene antisense to ATXN2, is associated with spinocerebellar ataxia type 2 and amyotrophic lateral sclerosis. Ann. Neurol. 80, 600–615 (2016).
Perry, R. B.-T., Hezroni, H., Goldrich, M. J. & Ulitsky, I. Regulation of neuroregeneration by long noncoding RNAs. Mol. Cell 72, 553–567 (2018).
Mehta, S. L., Kim, T. & Vemuganti, R. Long noncoding RNA FosDT promotes ischemic brain injury by interacting with REST-associated chromatin-modifying proteins. J. Neurosci. 35, 16443–16449 (2015).
Wu, Z. et al. LncRNA-N1LR enhances neuroprotection against ischemic stroke probably by inhibiting p53 phosphorylation. Mol. Neurobiol. 54, 7670–7685 (2017).
Xu, Q. et al. Long non-coding RNA C2dat1 regulates CaMKIIδ expression to promote neuronal survival through the NF-κB signaling pathway following cerebral ischemia. Cell Death Dis. 7, e2173 (2016).
Jung, E. S. et al. Genome-wide association analysis reveals the associations of NPHP4, TYW1-AUTS2 and SEMA6D for Behçet’s disease and HLA-B*46:01 for its intestinal involvement. Dig. Liver Dis. 56, 994–1001 (2024).
Kutlu, G., Semercioglu, S., Ucler, S., Erdal, A. & Inan, L. E. Epileptic seizures in neuro-Behcet disease: why some patients develop seizure and others not? Seizure 26, 32–35 (2015).
Zhou, Z.-D., Jankovic, J., Ashizawa, T. & Tan, E.-K. Neurodegenerative diseases associated with non-coding CGG tandem repeat expansions. Nat. Rev. Neurol. 18, 145–157 (2022).
Maharana, S. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018).
Biscarini, S. et al. Characterization of the lncRNA transcriptome in mESC-derived motor neurons: implications for FUS-ALS. Stem Cell Res. 27, 172–179 (2018).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Donnelly, C. J. et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 (2013).
Lagier-Tourenne, C. et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl Acad. Sci. USA 110, E4530–E4539 (2013).
Yokoshi, M. et al. Direct binding of Ataxin-2 to distinct elements in 3′ UTRs promotes mRNA stability and protein expression. Mol. Cell 55, 186–198 (2014).
Katsushima, K., Jallo, G., Eberhart, C. G. & Perera, R. J. Long non-coding RNAs in brain tumors. NAR Cancer 3, zcaa041 (2021).
Kim, S.-H., Lim, K.-H., Yang, S. & Joo, J.-Y. Long non-coding RNAs in brain tumors: roles and potential as therapeutic targets. J. Hematol. Oncol. 14, 77 (2021).
Chen, S. et al. Noncoding RNAs in pediatric brain tumors: molecular functions and pathological implications. Mol. Ther. Nucleic Acids 26, 417–431 (2021).
Wang, L. et al. CRISPR-Cas13d screens identify KILR, a breast cancer risk-associated lncRNA that regulates DNA replication and repair. Mol. Cancer 23, 101 (2024).
Mercer, T. R., Munro, T. & Mattick, J. S. The potential of long noncoding RNA therapies. Trends Pharmacol. Sci. 43, 269–280 (2022).
Egli, M. & Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 51, 2529–2573 (2023).
Jin, J. & Zhong, X.-B. ASO drug Qalsody (tofersen) targets amyotrophic lateral sclerosis. Trends Pharmacol. Sci. 44, 1043–1044 (2023).
Hagenacker, T. et al. Nusinersen in adults with 5q spinal muscular atrophy: a non-interventional, multicentre, observational cohort study. Lancet Neurol. 19, 317–325 (2020).
Liu, S. J. & Lim, D. A. Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep. 19, e46955 (2018).
Khorkova, O. et al. Long non-coding RNA-targeting therapeutics: discovery and development update. Expert Opin. Drug Discov. 18, 1011–1029 (2023).
Modarresi, F. et al. Knockdown of BACE1-AS nonprotein-coding transcript modulates beta-amyloid-related hippocampal neurogenesis. Int. J. Alzheimer’s Dis. 2011, 929042 (2011).
Meng, L. et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518, 409–412 (2015).
Hsiao, J. et al. Upregulation of haploinsufficient gene expression in the brain by targeting a long non-coding RNA improves seizure phenotype in a model of Dravet syndrome. eBioMedicine 9, 257–277 (2016).
Coan, M., Haefliger, S., Ounzain, S. & Johnson, R. Targeting and engineering long non-coding RNAs for cancer therapy. Nat. Rev. Genet. 25, 578–595 (2024).
Deguchi, S. et al. Investigator-initiated phase I trial of an oligonucleotide therapeutic targeting long noncoding RNA TUG 1 for recurrent glioblastoma. BMC Cancer 25, 251 (2025).
Neveu, M.-A. C. et al. Preclinical development of FTX-001: first in class inhibitor of the long non-coding RNA MALAT1. Cancer Res. 83, abstr. 450 (2023).
Dowdy, S. F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotech. 35, 222–229 (2017).
Roberts, T. C., Langer, R. & Wood, M. J. A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 19, 673–694 (2020).
Gonzales-Aloy, E., Ahmed-Cox, A., Tsoli, M., Ziegler, D. S. & Kavallaris, M. From cells to organoids: the evolution of blood-brain barrier technology for modelling drug delivery in brain cancer. Adv. Drug Deliv. Rev. 196, 114777 (2023).
Barker, S. J. et al. Targeting the transferrin receptor to transport antisense oligonucleotides across the mammalian blood-brain barrier. Sci. Transl. Med. 16, eadi2245 (2024).
Leitão, A. D. G. et al. Novel systemic delivery of a peptide-conjugated antisense oligonucleotide to reduce α-synuclein in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 186, 106285 (2023).
Yeoh, Y. Q. et al. Efficient systemic CNS delivery of a therapeutic antisense oligonucleotide with a blood-brain barrier-penetrating ApoE-derived peptide. Biomed. Pharmacother. 175, 116737 (2024).
Min, H. S. et al. Systemic brain delivery of antisense oligonucleotides across the blood–brain barrier with a glucose-coated polymeric nanocarrier. Angew. Chem. Int. Ed. 59, 8173–8180 (2020).
Porter, A. H., Johnson, N. A. & Tulchinsky, A. Y. A new mechanism for Mendelian dominance in regulatory genetic pathways: competitive binding by transcription factors. Genetics 205, 101–112 (2017).
Halow, J. M. et al. Tissue context determines the penetrance of regulatory DNA variation. Nat. Commun. 12, 2850 (2021).
Lappalainen, T., Li, Y. I., Ramachandran, S. & Gusev, A. Genetic and molecular architecture of complex traits. Cell 187, 1059–1075 (2024).
Wang, R. & Schlick, T. How large is the universe of RNA-like motifs? A clustering analysis of RNA graph motifs using topological descriptors. PLoS Comput. Biol. 21, e1013230 (2025).
Childs-Disney, J. L. et al. Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 21, 736–762 (2022).
Chen, S., Mao, Q., Cheng, H. & Tai, W. RNA-binding small molecules in drug discovery and delivery: an overview from fundamentals. J. Med. Chem. 67, 16002–16017 (2024).
Donlic, A. et al. Discovery of small molecule ligands for MALAT1 by tuning an RNA-binding scaffold. Angew. Chem. Int. Ed. 130, 13426–13431 (2018).
Abulwerdi, F. A. et al. Selective small-molecule targeting of a triple helix encoded by the long noncoding RNA, MALAT1. ACS Chem. Biol. 14, 223–235 (2019).
Davies, J. W. A., Bredy, T. W. & Marshall, P. R. Cutting-edge RNA technologies to advance the understanding of learning and memory. Neurobiol. Learn. Mem. 219, 108050 (2025). This review highlights tools that enable the study of learning and memory through cell sorting, RNA imaging, subcellular fractionation, laser microdissection, spatial transcriptomics, sequencing and manipulation.
Mardakheh, F. K. & Shechner, D. M. A molecular cartographer’s toolkit for mapping RNA’s uncharted realms. Cell Rep. 44, 115877 (2025).
Perez-Perri, J. I. et al. The RNA-binding protein landscapes differ between mammalian organs and cultured cells. Nat. Commun. 14, 2074 (2023).
Dodel, M. et al. TREX reveals proteins that bind to specific RNA regions in living cells. Nat. Methods 21, 423–434 (2024).
Tsue, A. F. et al. Multiomic characterization of RNA microenvironments by oligonucleotide-mediated proximity-interactome mapping. Nat. Methods 21, 2058–2071 (2024).
Biayna, J. & Dumbović, G. Decoding subcellular RNA localization one molecule at a time. Genome Biol. 26, 45 (2025).
Zollinger, D. R., Lingle, S. E., Sorg, K., Beechem, J. M. & Merritt, C. R. in In Situ Hybridization Protocols (eds Nielsen, B. S. & Jones, J.) 331–345 (Springer, 2020).
Goralski, T. M. et al. Spatial transcriptomics reveals molecular dysfunction associated with cortical Lewy pathology. Nat. Commun. 15, 2642 (2024).
Zhao, X., Lan, Y. & Chen, D. Exploring long non-coding RNA networks from single cell omics data. Comput. Struct. Biotech. J. 20, 4381–4389 (2022).
Goñi, E. et al. Uncovering functional lncRNAs by scRNA-seq with ELATUS. Nat. Commun. 15, 9709 (2024).
Gandal, M. J. et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018).
Mudge, J. M. et al. GENCODE 2025: reference gene annotation for human and mouse. Nucleic Acids Res. 53, D966–D975 (2024).
Zhao, L. et al. NONCODEV6: an updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 49, D165–D171 (2020).
Isik, S. & Unal, G. Open-source software for automated rodent behavioral analysis. Front. Neurosci. 17, 1149027 (2023).
Hsieh, C.-M., Hsu, C.-H., Chen, J.-K. & Liao, L.-D. AI-powered home cage system for real-time tracking and analysis of rodent behavior. iScience 27, 111223 (2024).
Isaacson, M. et al. MouseGoggles: an immersive virtual reality headset for mouse neuroscience and behavior. Nat. Methods 22, 380–385 (2025).
Judák, L. et al. Moculus: an immersive virtual reality system for mice incorporating stereo vision. Nat. Methods 22, 386–398 (2025).
Dumont, J. R., Salewski, R. & Beraldo, F. Critical mass: the rise of a touchscreen technology community for rodent cognitive testing. Genes Brain Behav. 20, e12650 (2021).
Palmer, D. et al. Touchscreen cognitive testing: cross-species translation and co-clinical trials in neurodegenerative and neuropsychiatric disease. Neurobiol. Learn. Mem. 182, 107443 (2021).
Davies, G. et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun. 9, 2098 (2018).
Zeng, B. et al. Multi-ancestry eQTL meta-analysis of human brain identifies candidate causal variants for brain-related traits. Nat. Genet. 54, 161–169 (2022).
Zhong, W. et al. Understanding the function of regulatory DNA interactions in the interpretation of non-coding GWAS variants. Front. Cell Dev. Biol. 10, 957292 (2022).
Savage, J. E. et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 50, 912–919 (2018).
Hill, W. D. et al. A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol. Psych. 24, 169–181 (2019).
Roussos, P. et al. A role for noncoding variation in schizophrenia. Cell Rep. 9, 1417–1429 (2014).
Bartonicek, N. et al. Intergenic disease-associated regions are abundant in novel transcripts. Genome Biol. 18, 241 (2017).
Hardwick, S. A. et al. Targeted, high-resolution RNA sequencing of non-coding genomic regions associated with neuropsychiatric functions. Front. Genet. 10, 309 (2019).
St. Laurent, G., Vyatkin, Y. & Kapranov, P. Dark matter RNA illuminates the puzzle of genome-wide association studies. BMC Med. 12, 97 (2014).
Morselli Gysi, D. & Barabási, A.-L. Noncoding RNAs improve the predictive power of network medicine. Proc. Natl Acad. Sci. USA 120, e2301342120 (2023).
GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
de Goede, O. M. et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell 184, 2633–2648 (2021).
López Soriano, V. et al. Multi-omics analysis in human retina uncovers ultraconserved cis-regulatory elements at rare eye disease loci. Nat. Commun. 15, 1600 (2024).
Deng, Y. et al. Loss of LAMP5 interneurons drives neuronal network dysfunction in Alzheimer’s disease. Acta Neuropathol. 144, 637–650 (2022).
Durand-de Cuttoli, R. et al. Change-of-mind neuroeconomic decision-making is modulated by LINC00473 in medial prefrontal cortex in a sex-dependent manner. Sci. Adv. 11, eadr3228 (2025).
Bernard, D. et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093 (2010).
Carrieri, C. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 (2012).
Faghihi, M. A. et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730 (2008).
Perry, R. B.-T., Tsoory, M., Tolmasov, M. & Ulitsky, I. Silc1 long noncoding RNA is an immediate-early gene promoting efficient memory formation. Cell Rep. 42, 113168 (2023).
Tan, M. C. et al. The activity-induced long non-coding RNA Meg3 modulates AMPA receptor surface expression in primary cortical neurons. Front. Cell. Neurosci. 11, 124 (2017).
Balusu, S. et al. MEG3 activates necroptosis in human neuron xenografts modeling Alzheimer’s disease. Science 381, 1176–1182 (2023).
Klymenko, A. et al. Downregulation of lncRNAs Gomafu, NONMMUT033604.2, and NONMMUT064397.2 in the hippocampus of mice with model of post-traumatic stress disorder. World J. Biol. Psych. 25, 283–290 (2024).
Luo, J. et al. LncRNAs: architectural scaffolds or more potential roles in phase separation. Front. Genet. 12, 369 (2021).
Dunker, A. K., Bondos, S. E., Huang, F. & Oldfield, C. J. Intrinsically disordered proteins and multicellular organisms. Semin. Cell Dev. Biol. 37, 44–55 (2015).
Yruela, I., Oldfield, C. J., Niklas, K. J. & Dunker, A. K. Evidence for a strong correlation between transcription factor protein disorder and organismic complexity. Genome Biol. Evol. 9, 1248–1265 (2017).
Niklas, K. J., Dunker, A. K. & Yruela, I. The evolutionary origins of cell type diversification and the role of intrinsically disordered proteins. J. Exp. Bot. 69, 1437–1446 (2018).
Jonas, F., Navon, Y. & Barkai, N. Intrinsically disordered regions as facilitators of the transcription factor target search. Nat. Rev. Genet. 26, 424–435 (2025).
Bondos, S. E., Dunker, A. K. & Uversky, V. N. On the roles of intrinsically disordered proteins and regions in cell communication and signaling. Cell Commun. Signal. 19, 88 (2021).
Afanasyeva, A., Bockwoldt, M., Cooney, C. R., Heiland, I. & Gossmann, T. I. Human long intrinsically disordered protein regions are frequent targets of positive selection. Genome Res. 28, 975–982 (2018).
Hentze, M. W., Castello, A., Schwarzl, T. & Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 (2018). This article reveals that RNA-binding domains are widespread in eukaryotic proteins, including in metabolic proteins.
Bah, A. & Forman-Kay, J. D. Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 291, 6696–6705 (2016).
Darling, A. L. & Uversky, V. N. Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front. Genet. 9, 158 (2018).
Wesseling, H. et al. Tau PTM profiles identify patient heterogeneity and stages of Alzheimer’s disease. Cell 183, 1699–1713 (2020).
Lepack, A. E. et al. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 368, 197–201 (2020). This paper shows that neurotransmitters also have a role in epigenetic processes.
Uversky, V. N. Intrinsically disordered proteins and their (disordered) proteomes in neurodegenerative disorders. Front. Aging Neurosci. 7, 18 (2015).
Zbinden, A., Pérez-Berlanga, M., De Rossi, P. & Polymenidou, M. Phase separation and neurodegenerative diseases: a disturbance in the force. Dev. Cell 55, 45–68 (2020).
Malik, I., Kelley, C. P., Wang, E. T. & Todd, P. K. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 22, 589–607 (2021).
Tsang, B., Pritišanac, I., Scherer, S. W., Moses, A. M. & Forman-Kay, J. D. Phase separation as a missing mechanism for interpretation of disease mutations. Cell 183, 1742–1756 (2020).
Ahmed, R. & Forman-Kay, J. D. Aberrant phase separation: linking IDR mutations to disease. Cell Res. 33, 583–584 (2023).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Jain, A. & Vale, R. D. RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017).
Basu, S. et al. Unblending of transcriptional condensates in human repeat expansion disease. Cell 181, 1062–1079 (2020).
Das, T. et al. Tunable metastability of condensates reconciles their dual roles in amyloid fibril formation. Mol. Cell 85, 2230–2245.e7 (2025).
Fernandopulle, M. S., Lippincott-Schwartz, J. & Ward, M. E. RNA transport and local translation in neurodevelopmental and neurodegenerative disease. Nat. Neurosci. 24, 622–632 (2021).
Bauer, K. E., de Queiroz, B. R., Kiebler, M. A. & Besse, F. RNA granules in neuronal plasticity and disease. Trends Neurosci. 46, 525–538 (2023).
Kiebler, M. A. & Bauer, K. E. RNA granules in flux: dynamics to balance physiology and pathology. Nat. Rev. Neurosci. 25, 711–725 (2024).
Pechstein, A. et al. Vesicle clustering in a living synapse depends on a synapsin region that mediates phase separation. Cell Rep. 30, 2594–2602 (2020).
Chen, X., Wu, X., Wu, H. & Zhang, M. Phase separation at the synapse. Nat. Neurosci. 23, 301–310 (2020). This is one of many papers showing that synaptic vesicles are phase-separated domains, which is consistent with a role for lncRNAs in their formation and function.
Sansevrino, R., Hoffmann, C. & Milovanovic, D. Condensate biology of synaptic vesicle clusters. Trends Neurosci. 46, 293–306 (2023).
Jin, G. et al. The liprin-α/RIM complex regulates the dynamic assembly of presynaptic active zones via liquid–liquid phase separation. PLoS Biol. 23, e3002817 (2025).
Lützkendorf, J. & Sigrist, S. J. Condensate dynamics at the synapse: phase separation tunes presynaptic function. PLoS Biol. 23, e3003201 (2025).
Bakthavachalu, B. et al. RNP-granule assembly via ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98, 754–766 (2018).
Novoa, E. M., Mason, C. E. & Mattick, J. S. Charting the unknown epitranscriptome. Nat. Rev. Mol. Cell Biol. 18, 339–340 (2017).
Jonkhout, N. et al. The RNA modification landscape in human disease. RNA 23, 1754–1769 (2017).
Flores, J. V. et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Rep. 8, 112–124 (2017).
Ma, C. et al. RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19, 68 (2018).
Merkurjev, D. et al. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014 (2018).
Engel, M. et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron 99, 389–403 (2018).
Widagdo, J. et al. Experience-dependent accumulation of N-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36, 6771–6777 (2016). This paper supports a role for RNA modifications in cognitive function.
Mladenova, D. et al. Adar3 is involved in learning and memory in mice. Front. Neurosci. 12, 243 (2018).
Begik, O., Mattick, J. S. & Novoa, E. M. Exploring the epitranscriptome by native RNA sequencing. RNA 28, 1430–1439 (2022).
Freude, K. et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am. J. Hum. Genet. 75, 305–309 (2004).
Khan, M. A. et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90, 856–863 (2012).
Davarniya, B. et al. The role of a novel TRMT1 gene mutation and rare GRM1 gene defect in intellectual disability in two Azeri families. PLoS ONE10, e0129631 (2015).
Zhang, K. et al. An intellectual disability-associated missense variant in TRMT1 impairs tRNA modification and reconstitution of enzymatic activity. Hum. Mutat. 41, 600–607 (2020).
Vauti, F. et al. The mouse Trm1-like gene is expressed in neural tissues and plays a role in motor coordination and exploratory behaviour. Gene 389, 174–185 (2007).
Jonkhout, N. et al. Subcellular relocalization and nuclear redistribution of the RNA methyltransferases TRMT1 and TRMT1L upon neuronal activation. RNA Biol. 18, 1905–1919 (2021). This paper shows specific subcellular localizations of RNA modification enzymes in neurons and their relocalization upon neuronal stimulation, indicative of the exquisite transactions that must be taking place.
Yang, L. et al. Emerging role of RNA modification and long noncoding RNA interaction in cancer. Cancer Gene Ther. 31, 816–830 (2024).
Acknowledgements
This work was supported by UNSW Sydney (grant RG193211) and the Australian Research Council (grants DP210103233 and DP210101957). The authors thank K. Clemens (UNSW Sydney) for the helpful comments on the manuscript. Figures were created with BioRender (https://www.biorender.com).
Author information
Authors and Affiliations
Contributions
S.A. and M.J.C. assembled and curated the lists of lncRNAs and their reported functions, study models, expression patterns and phenotypic effects of knockdown or ectopic expression. S.A. also drafted the figures. J.S.M. wrote the first draft of the article and managed the revisions. L.M.I. contributed to the section ‘LncRNAs in neurological disorders’. All authors edited the article and the revised versions and approved their submission.
Corresponding author
Ethics declarations
Competing interests
L.M.I. is the Founder and Chief Medical Officer of Celosia Therapeutics Pty Ltd.
Peer review
Peer review information
Nature Reviews Neuroscience thanks Yue Feng, Gerhard Schratt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Antisense lncRNAs
-
RNAs that are transcribed from the strand opposite a protein-coding gene and overlap the body of the gene (in part of full) or its promoter.
- Enhancers
-
Genetic loci that activate the spatiotemporal patterns of the expression of genes in their vicinity, independent of position, orientation or (to a limited extent) distance.
- Genome editing
-
A technique that allows precise removal, replacement or addition of specific sequences in the genome, most commonly using engineered CRISPR–Cas systems.
- Genome-wide association study (GWAS) haplotype blocks
-
Genomic segments subject to limited internal recombination, allelic variants of which can be tagged by sentinel single-nucleotide polymorphisms and linked to various human complex traits and disorders in large-scale genome wide association studies.
- MicroRNAs
-
Small 21–25-nt non-coding RNAs that regulate mRNA translation and half-life by recruiting the RNA-induced silencing complex.
- Non-coding RNAs
-
RNAs that are not translated into protein but have functions in gene regulation.
- Proximity labelling
-
A biochemical technique that uses engineered enzymes to covalently tag proteins or RNAs near a protein of interest, enabling identification of interaction partners with spatial precision.
- Pseudogenes
-
Genomic DNA sequences derived from protein-coding genes, derived by retrotransposition of processed mRNAs or segmental duplication, which lack protein-coding capacity owing to insertions, deletions or premature stop codons.
- RNA transport granules
-
Motile phase-separated condensates of RNAs and proteins that mediate long-distance RNA trafficking along axons and dendrites.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Altaf, S., Cummins, M.J., Ittner, L.M. et al. The emerging roles of long non-coding RNAs in the nervous system. Nat. Rev. Neurosci. 26, 661–676 (2025). https://doi.org/10.1038/s41583-025-00960-z
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41583-025-00960-z
This article is cited by
-
The emerging role of epigenetic regulation in pain sensitization associated with headache disorders
The Journal of Headache and Pain (2025)