Abstract
Artificial intelligence has created tremendous advances for many scientific and engineering applications. In this Review, we synthesize recent advances in joint brain–behaviour modelling of neural and behavioural data, with a focus on methodological innovations, scientific and technical motivations, and key areas for future innovation. We discuss how these tools reveal the shared structure between the brain and behaviour and how they can be used for both science and engineering aims. We highlight how three broad classes with differing aims — discriminative, generative and contrastive — are shaping joint modelling approaches. We also discuss recent advances in behavioural analysis approaches, including pose estimation, hierarchical behaviour analysis and multimodal-language models, which could influence the next generation of joint models. Finally, we argue that considering not only the performance of models but also their trustworthiness and interpretability metrics can help to advance the development of joint modelling approaches.
Similar content being viewed by others
Introduction
Understanding how the brain gives rise to complex behaviour remains one of the central challenges in neuroscience. Although decades of research have elucidated the neural mechanisms underlying simple sensory or motor tasks, a mechanistic understanding of higher-order behaviours, such as decision-making, social interaction and cognitive flexibility, remains elusive. Progress in this domain is critically dependent on our ability to link brain activity with behaviour at appropriate levels of abstraction and resolution1,2,3. Joint brain–behaviour modelling has been a key methodological advance towards achieving that goal.
Recent years have seen major advances in both neural recording technologies and behavioural measurement tools4,5,6,7. On the neural side, large-scale electrophysiology, calcium imaging and neuromodulatory tagging enable the simultaneous recording of activity from hundreds to thousands of neurons across multiple brain regions5,8,9. On the behavioural side, high-resolution video, inertial sensors and pose estimation techniques have made it possible to capture fine-grained behavioural dynamics over time2,7,10,11,12,13. These parallel advances open the door to a deeper understanding of how distributed neural populations coordinate to drive complex behaviours, but only if they are integrated analytically.
Artificial intelligence (AI), which encompasses modern machine learning, deep learning and agent-based systems, has created tremendous advances for many scientific applications, ranging from protein design14 to weather prediction15. Naturally, AI also has had a tremendous impact in neuroscience on joint modelling approaches, which provide a statistical and computational framework to bridge neural and behavioural data. Rather than analysing each domain in isolation, joint models capture the shared structure between neural dynamics and behavioural outputs, enabling researchers to test hypotheses about how neural data are related to behaviour and vice versa (see refs. 16,17 for excellent probabilistic neural modelling reviews).
In this Review, we survey recent progress in joint modelling of neural and behavioural data, with a focus on methodological innovations, scientific and engineering motivations, and key areas for future innovation. We begin by giving some background on advances in AI that are relevant for understanding neural, behavioural and joint modelling approaches. We then survey the main optimization approaches relevant for joint modelling — discriminative, generative and contrastive — along with their limitations and advantages. We next discuss how these tools reveal the shared structure between neural activity and behaviour and how they can be used for both scientific and engineering aims. Then, we describe recent advances in behavioural analysis approaches, including hierarchical behaviour analysis, which could influence the next generation of joint models. Finally, we argue how considering not only the performance of models but also metrics of their trustworthiness and interpretability can help to advance the development of joint modelling approaches.
Principles of deep learning models
Fundamentally, the goal of AI models often amounts to solving challenging perception and decision-making problems. For instance, one needs to decide based on the recorded audio signal whether a rat is emitting an ultrasonic vocalization18 or based on a video whether the rat is doing unsolicited jumps (Freudensprünge)19. Experts can readily score such events, and it should be no surprise that AI systems are also increasingly capable of doing so. In broad strokes, these perception problems can now be solved with AI. Here, it is also worthwhile to remember that AI systems at times solve perception problems with algorithms that are at least loosely inspired by the brain20,21. In this section, we look more closely at how these AI systems achieve such perceptual capabilities — focusing on machine learning and deep learning fundamentals that underlie their success. By briefly examining how these methods operate and differ, we can better appreciate both their power and their limitations for joint modelling of neural data and behaviour.
Machine learning systems consist of four key components that work together to solve problems: a data set, a model, a loss function and an optimization algorithm22,23,24. The data set defines the input–output relationships that the model should learn; for instance, for ultrasonic vocalization identification, the system must predict a binary output (no call versus call) from a particular audio waveform input. The model serves as the mathematical framework that transforms these inputs into outputs through adjustable internal parameters. The loss function measures the quality of the model’s predictions by comparing them with the ground truth data, providing a numerical score that assesses performance. Loss functions quantify prediction error and are closely related to objective functions — the general term for any function being optimized (whether minimized or maximized). Finally, the optimization algorithm iteratively updates the model’s parameters to minimize this loss, effectively steering the model towards better performance. The specific choices made about these four components directly influence both the possible performance and the robustness of the overall machine learning system. Technically, this is the definition of supervised learning systems when the data have labels, namely, where input–output pairs are given. We later discuss self-supervised learning, which learns from unlabelled data by creating supervisory signals from the data’s own structure. This self-supervised paradigm lies at the heart of innovations for joint brain–behaviour modelling.
Before the advent of deep learning, classic (supervised) machine learning used domain-specific feature engineering (via a fixed encoder) followed by trainable classification (via a decoder). For ultrasonic vocalization processing, raw waveforms could be transformed via auditory filter banks into statistical descriptors (akin to what the cochlea does). In this case, those filter banks are the encoder. They extract features from the raw waveforms and these features are fed into a classifier (decoder) to predict calls. Only the decoder is trained whereas the encoder remains fixed, reflecting historical constraints where domain knowledge in the encoder compensated for limited learning capacity in the decoder.
Deep learning revolutionized this approach by making both the encoder and the decoder (alternatively called the backbone and output heads) learnable components implemented as deep neural networks. These networks consist of multiple layers of differentiable, non-linear transformations that are optimized together. Unlike classic approaches that rely on handcrafted features, deep neural networks optimize the feature representation directly for the task at hand, learning which aspects of the input are most relevant22,23. Neural networks, particularly deep architectures, excel in extracting hierarchical features that progress from simple local patterns to complex global structures. Given sufficient training data, this end-to-end learning yields superior performance and robustness. Typical model architectures are multilayer perceptrons (MLPs) (Fig. 1a), convolutional neural networks (CNNs) (Fig. 1b), recurrent neural networks (RNNs) (Fig. 1c), transformers (Fig. 1d) or state-space models (Fig. 1e). Although these architectures differ in structure, they all function as universal approximators capable of learning complex mappings when provided with enough capacity (model size; the number of adjustable internal parameters) and data25. However, the choice of architecture for both the encoder and the decoder may substantially impact both data efficiency and final performance24.
a, Multilayer perceptrons (MLPs) are neural networks composed of fully connected layers, where each neuron receives weighted input from every neuron in the preceding layer. This dense connectivity allows MLPs to learn complex non-linear mappings between inputs and outputs, although at the cost of a large number of parameters. b, Convolutional neural networks (CNNs) process grid-structured data such as images by applying learnable filters across spatial dimensions. In this toy example, three initial convolutions with weight sharing create feature maps, which are then downsampled via pooling to reduce spatial dimensions while retaining important features. Additional convolutions follow and, finally, fully connected layers predict outputs. CNNs exploit weight sharing and hierarchical feature extraction. In vision tasks, it is well known that they progressively build from edge detectors to complex object representations. c, Recurrent neural networks (RNNs) such as gated recurrent unit networks (GRUs) or long short-term memory networks (LSTMs) process sequential data by maintaining a hidden state h(t) that evolves over time t, passing information from one time step to the next via a recurrent connection that combines h(t − 1) and x(t). This recurrent connection acts as the network’s memory, allowing RNNs to capture temporal dependencies and patterns in sequential input data x(t) to create the output y(t). They can struggle with long-range temporal dependencies due to vanishing or exploding gradients. d, Transformers have revolutionized sequence modelling by replacing recurrent connections with self-attention mechanisms. These mechanisms compute relationships between all positions in a sequence simultaneously, enabling the capture of long-range temporal dependencies while maintaining computational parallelizability — a key advantage over sequential architectures. Transformers process input tokens (embeddings (emb)) combined with positional embeddings (post enc) through layers containing multi-head attention, MLPs and skip connections. Skip connections (also called residual connections) bypass intermediate layers (here, attention and MLP). ‘Add and norm’ blocks implement these skip connections: ‘add’ sums the input with the layer output (residual connection) whereas ‘norm’ applies layer normalization, together improving gradient flow and training stability35. e, State-space models provide an alternative approach to sequence processing by modelling data as continuous-time dynamical systems also with hidden states (h(t)). Through learned state transitions and efficient discretization schemes, state-space models can handle extremely long sequences with linear computational complexity as indicated by the state equations, making them particularly attractive for tasks requiring long-context understanding.
The loss function shapes what the model learns by defining success. In supervised learning, where there are labelled examples, the loss function typically measures the prediction error, such as using cross-entropy for classification tasks or the mean squared error (MSE) for regression (Box 1). The optimization process that minimizes the loss generally employs gradient-based methods24. Collectively, this framework of data set, model, loss and optimization provides a unified lens for understanding all types of machine learning systems, establishing the vocabulary that we use throughout our Review.
What is brain–behaviour modelling, and what is the goal?
Understanding how neural activity gives rise to hierarchical behaviour requires integrative modelling approaches that can extract structure from high-dimensional, heterogeneous data modalities. Overall, one is interested in modelling the joint distribution P(behaviour, neural data), which can be achieved in numerous different ways and generally falls into four classes. Decoding models study how behaviour depends on neural data, P(behaviour | neural data). Encoding models instead study how neural data depend on behaviour (or more sensory input), P(neural data | behaviour). Latent models capture P(neural data) via self-supervised learning using generative or contrastive approaches, learning latent variables z that are then related to behavioural data. Latent variables are unobserved quantities that must be inferred from observed data and typically represent abstract features that capture underlying structure in high-dimensional observations. For example, whereas thermometers appear to measure it directly, temperature is fundamentally a latent variable — a statistical property of particle energy distributions that we infer from microscopic states. Similarly, in neural–behavioural modelling, latent variables represent aggregate properties of high-dimensional neural activity that we infer from observable spike trains and behaviour. Finally, joint models directly model the joint distribution of behaviour and neural data, P(behaviour, neural data). These different approaches immediately raise the question of which modelling approach is best suited for a given scientific or engineering goal.
From an engineering perspective, one may aim to build brain–machine interfaces (BMIs), where high performance in behavioural decoding and real-time execution are paramount. By contrast, a scientific goal may involve constructing a mechanistic model that captures the computational principles and dynamical processes underlying neural function — analogous to a digital twin in engineering, but focused on biological principles rather than exact replication. Such models should not only reproduce observed neural–behavioural relationships but also enable discovery of new principles through simulation, perturbation and hypothesis generation. Alternatively, the goal may be to test specific hypotheses about neural representations, necessitating interpretable latent variables that can be experimentally validated or falsified. A fourth objective involves exploratory discovery — using these methods to uncover novel patterns, cell types or computational motifs that were not previously known or hypothesized.
Each of these goals imposes different modelling requirements and evaluation criteria. Crucially, the answer cannot rely solely on decoding performance metrics such as spike prediction accuracy or behavioural reconstruction, as these metrics conflate fundamentally different objectives. A model achieving 99% decoding accuracy might use biologically implausible transformations that provide little insight into neural computation, making it excellent for BMI applications but unsuitable for mechanistic understanding. Therefore, articulating the scientific intent — whether engineering performance, mechanistic insight, hypothesis testing or open-ended discovery — is essential for guiding model selection, development and interpretation.
The diversity of scientific and engineering goals has naturally led to the development of these multiple modelling paradigms. Notably, whether one creates decoding, encoding, latent or joint models, broadly speaking there are three computational objectives (Fig. 2). Discriminative objectives for decoding are those that aim to predict behaviour (for example, spikes in, decode behaviour); however, we note that encoding models also use discriminative approaches and map from behaviour and/or stimuli to spikes3,26. Generative objectives for reconstruction are those that aim to reconstruct input data from learned latent representations (for example, spikes or behaviour in, predict spikes or behaviour). Contrastive objectives for encoding and joint modelling are those that aim to encode without reconstruction (for example, spikes and behaviour in, learn latents via representation learning). Although each of these three approaches can serve either a specific goal or multiple goals, they all flourish due to complementary trade-offs: discriminative models provide computational efficiency for targeted predictions; generative models enable sampling and uncertainty quantification; and contrastive methods leverage unlabelled data to discover representations that generalize across contexts. In the following sections, we examine each paradigm in detail, highlighting representative methods and their applications to neural–behavioural data.
For all approaches, encoders and decoders can comprise different architectures (Fig. 1) and these learned models (encoders or decoders) can then be leveraged in downstream tasks. Although we focus on spikes as the primary input for illustration, other types of neural recordings (such as calcium imaging, local field potentials or functional MRI) can also be used with these approaches. a, Discriminative approaches use input spikes to decode behaviour using supervised losses such as the mean squared error (MSE) for continuous variables and cross-entropy for discrete variables. During training, predicted behaviour is compared with ground truth behaviour to compute the loss and update model parameters. At inference time, the trained model outputs predict behaviour without requiring ground truth. b, In generative approaches, a model comprising an encoder and a decoder learns to generate spike data from latents. The encoder maps input spikes to latent representations, whereas the decoder reconstructs spikes from these latent codes. Reconstruction losses such as the negative log likelihood (NLL) or MSE compare predicted with ground truth spikes. In variational autoencoder (VAEs), training optimizes the evidence lower bound, which combines reconstruction loss with regularization on the latent distribution (Box 1). c, Contrastive approaches use input spikes, optionally with auxiliary variables (such as behaviour labels), to learn latent representations through contrastive learning. This approach achieves representation learning without explicit reconstruction (decoding). Namely, learning is based on attraction and repulsion dynamics: similar samples (positive pairs) are pulled together whereas dissimilar samples (negative pairs) are pushed apart in the latent representation. CNN, convolutional neural network; InfoNCE, information noise-contrastive estimation; NCE, noise-contrastive estimation; RNN, recurrent neural network.
Discriminative models: directly decoding behaviour from neural data
Decoding is a long-standing task in neuroscience, beginning with classical approaches such as population vectors and Kalman filters (reviewed elsewhere3). These methods established the basic framework for mapping high-dimensional neural activity to low-dimensional behavioural variables, which allows for both understanding the information present in a population of neurons and for engineering BMIs. As the field progressed, machine learning techniques such as support vector machines27 and decision trees were adopted to improve decoding performance. Today, in terms of performance, these have largely been superseded by deep learning models, including transformer-based architectures28,29 (Fig. 1d).
These modern decoding models are typically supervised, using behaviour directly as the target in the loss function, most often with the MSE (Box 1). Recent benchmarking efforts30,31 have formalized this, focusing on the accuracy of behavioural decoding (and the prediction of spikes) (see the section ‘Generative models: learning to predict spike trains via reconstruction’) as the key measure of success. Note that ‘behaviour’ is typically a discrete or continuous 2D variable, such as the velocity of the hand or 2D position of a cursor on a screen, but in the following we discuss how new approaches to measuring behaviour could change the nature of this decoding goal (see the section ‘Behavioural analysis for neuroscience’).
Indeed, transformer architectures are making impressive gains for decoding28,29,32,33. Their ability to flexibly model long-range dependencies and multimodal inputs has enabled state-of-the-art performance in behavioural decoding in comparison with supervised MLPs and RNNs (Fig. 1). One key advantage of transformers is their scalability: their architecture enables parallel computation, efficient use of large data sets and improved performance with increasing model size. Although attention operations are computationally expensive, self-attention can flexibly integrate contextual cues such as trial structure, sensory stimuli or task rules (Figs. 1d and 2a). This makes them especially suitable for data sets with complex temporal structure. Notably, tokenization of the spikes and leveraging positional embedding makes combining multi-session, multi-animal data more feasible. Newer scalable transformers such as Perceiver I/O offer greater flexibility and predictive power34. This enables fine-tuning and generalization to held-out data sets, paving the way for better foundation models29 (Box 2).
Yet there are also clear trade-offs in the complexity and speed of using large transformer models35,36, which has limitations for the deployment on devices37 and, practically speaking, the weeks of compute required for training29 can make this approach not viable for many laboratories. Therefore, although many new powerful approaches have been proposed in terms of decoding performance, there are ongoing efforts to build lighter-weight unified models that perform equally well even with smaller RNNs or MLPs38. For example, Sani et al.38 developed powerful lightweight models to extract task-relevant and task-irrelevant latent dynamics.
Generative models: learning to predict spike trains via reconstruction
Reconstruction-based approaches represent a powerful paradigm for learning latent representations of neural data without requiring labelled examples. Variational autoencoders (VAEs) are particularly well suited for neural data analysis because they learn probabilistic mappings between high-dimensional observations (x) and a (typically lower-dimensional) latent variable (z)39,40. A VAE consists of an encoder qϕ(z|x) (recognition model) that approximates the true but intractable posterior distribution (the probability of latent variables given observed data, p(z|x)), and a decoder (pθ(x|z)) that reconstructs the observations (data) from these latents by optimizing the data likelihood (Fig. 2b and Box 1). Unlike deterministic autoencoders, which map each input to a single point in latent space, VAEs learn probability distributions over latent representations, and thus model uncertainty in both the latent variables and the reconstruction process. This makes them especially valuable for capturing the inherent variability of neural data. We emphasize that VAEs are generative models, and the encoder is both a technical solution to learn the generative model and also a way to infer latent variables from data. VAEs are used in both of these ways in the literature (for example, see latent factor analysis via dynamical systems (LFADS) below). Importantly, the generative nature of VAEs also enables sampling novel neural patterns and quantifying uncertainty in latent variables (also called latent representations), which is essential for understanding the probabilistic structure underlying neural population activity.
In neuroscience, LFADS pioneered the application of VAEs by combining them with RNNs to model neural activity as a dynamical system41,42. LFADS can infer both trial-specific latent trajectories and putative inputs to the neural dynamics one is modelling. These learned latents and these input dynamics can then be related to behavioural and other experimental variables. To give some concrete examples, the learned representations have proven effective in decoding primate hand movements from the motor cortex and in detecting perturbations41. One can also learn models of neural dynamics across multiple experimental sessions (stitching) and use the generative nature of LFADS for sampling synthetic data41,42.
Whereas LFADS assumes continuous latent dynamics, switching linear dynamical systems (SLDS) takes a different approach by modelling neural activity governed by discrete state transitions. SLDS extends traditional state-space frameworks (Fig. 1e) by allowing the system to transition between multiple latent dynamical regimes over time. In neuroscience, these models have been used to flexibly capture non-stationary neural population dynamics. By inferring a sequence of discrete states from neural data, with each regime governed by distinct dynamics, SLDS models can reveal behaviourally relevant brain state switches, cognitive modes or neural circuit configurations43,44,45,46. For data from multiple individuals, it can be important to consider families of dynamical systems that share some parameters across individuals such as multi-task dynamical systems47. In general, their strength lies in their interpretability of the latent states that can demarcate transitions in neural dynamics. For example, one could use the resulting model to predict a context change or behavioural action switch from neural dynamics.
The reconstruction-based approach that defines VAEs is both their strength and their fundamental limitation. These methods optimize in raw data space where natural metrics (such as pixel distance or Poisson loss) may not capture meaningful similarity in the underlying (latent) structure48. For instance, neurons deviate from Poisson statistics but are commonly modelled in this way. The reconstruction requirement forces a trade-off between capturing input fidelity and learning task-relevant latent representations: capacity spent on high-fidelity reconstruction may not be available for capturing task-relevant latent structure. There is no guarantee that minimizing the reconstruction error will yield representations that are optimal for understanding neural–behavioural relationships. This misalignment between the metric used for reconstruction and the actual latent representation is a key challenge: the natural metric for reconstruction may not align with the meaningful structure in the data. This reconstruction challenge is evident in vision applications, where standard VAE objectives often produce blurry reconstructions — the model optimizes what one measures (pixel similarity) rather than what one cares about (perceptual quality). This motivated the development of more sophisticated generative approaches with diffusion models3,49. Recent work also leverages diffusion models and state-space models (Fig. 1e) to more realistically generate neural activity50. Another important limitation is that VAEs suffer from not producing consistent results (Box 1).
Contrastive models: learning latents without reconstructing data
Contrastive learning sidesteps the reconstruction dilemma. Instead of asking ‘how do we generate this neural pattern?’, contrastive methods ask ‘what makes this neural pattern similar to or different from other patterns?’. This reframing eliminates the need to specify spike-level (Poisson loss) or pixel-level (pixel distance) similarity metrics, allowing the model to focus on discovering native relationships in the data51,52,53,54,55. Contrastive learning learns latent representations by maximizing agreement between related samples (positive pairs) while minimizing agreement between unrelated samples (negative pairs), without requiring supervised (behavioural) labels or input reconstruction (such as spike reconstruction) (Fig. 2c). Crucially, such models avoid imposing strong generative assumptions or supervised targets, which may bias or constrain the learned representation.
A method called CEBRA has pioneered this approach for continuous and discrete time-series data, particularly neural data54,55. CEBRA operates by pulling positive pairs closer in latent space while pushing negative pairs, typically using objectives such as information noise-contrastive estimation (InfoNCE)51,56 (Box 2). For neural data, temporal proximity can serve as a natural basis for defining positive and negative pairs — neural patterns occurring within short time windows are treated as related (positive), whereas patterns separated by longer intervals serve as unrelated (negative) samples54. This self-supervised objective promotes embeddings that reflect the intrinsic temporal structure of neural data, capturing cognitive states and behavioural dynamics without requiring explicit labels or a supervised loss function. A core flexibility of the contrastive approach lies in how the positive and negative pairs are defined. A current limitation is that the time window is a tunable parameter but restricted to a single timescale; future efforts should allow for hierarchical time bins. Importantly, this approach naturally can be extended to joint modelling.
Joint models for inferring latent dynamics via representation learning
Indeed, in addition to using time-aware contrastive loss, CEBRA54 can also use time-aware plus auxiliary variables (labels) to guide which neural samples to attract together in the latent space. This use of labels allows for joint modelling of behavioural–neural data in a hypothesis-guided manner. Because the positive pairs can be crafted from the auxiliary variables (for example, behaviour), this explicitly allows for testing which behavioural labels extract meaningful latents from the neural space. For instance, if ‘space’ is hypothesized to be encoded in a given neural population, close spatial distances of the animal can be used to sample the positive pairs, and far spatial distances for negative pairs. If this relationship between space and the neural data does not exist, Schneider et al. showed both empirically and theoretically that this creates an unsolvable optimization problem — the model cannot simultaneously satisfy the contrastive constraints — and the embedding collapses to a trivial solution on the hypersphere (a diffuse cloud distributed on it)54,55. Note that auxiliary variables can also be derived from other modalities such as video embeddings54.
Such joint modelling with contrastive learning generalized theory from non-linear independent component analysis to ensure identifiability of the model (Box 1). Specifically, if two models f and \(\mathop{f}\limits^{ \sim }\) trained on the same data yield the same conditional distributions over sample pairs (for example, via InfoNCE loss) (Box 1), then their embeddings are linearly related — that is, a transformation L exists such that \(\mathop{f}\limits^{ \sim }(x)=Lf(x)\) for all x in the data set. This identifiability ensures that downstream tasks relying on these embeddings will behave consistently across (different) model instantiations. This consistency enables robust use in downstream tasks such as decoding or topological data analysis, and facilitates cross-participant or cross-modality alignment. As it only requires that latent variables vary sufficiently over time, CEBRA provides a flexible framework for analysing complex neural data (whether spikes or imaging) or behaviour, and can recover latent trajectories aligned with meaningful experimental variables under mild assumptions.
New work has extended this framework to include explicit temporal dynamics priors. Dynamic contrastive learning has incorporated explicit modelling of the SLDS to extract hypothesis-guided dynamical systems from neural data57. MARBLE also leverages contrastive learning, but first preprocesses the neural activity via geometric deep learning approaches into manifold embeddings58. By doing so, they implicitly incorporate similarity through the similarity of spiking patterns over time. The limitation is that enforcing a specific geometry may restrict flexibility in capturing latent neural dynamics that do not conform to the assumed manifold structure.
Another key feature of these approaches is the identifiability of the models (Box 1). As we discussed in this section, contrastive learning with auxiliary variables can uniquely recover models when networks are bijective under noise-contrastive estimation (NCE) loss, and with InfoNCE loss the bijectivity assumption is sometimes unnecessary53,54,55,57. Notably, identifiability can also be achieved with VAEs (under more specific generative model assumptions). For the relevant neuroscientific case of Poisson noise, this was carried out in PI-VAE59. PI-VAE built on advances in identifiable VAEs60 (Box 1) to develop a method that outperformed LFADS, VAEs and pfLDS44 in predicting the latent variables in the underlying data, namely the position of a rat navigating on a linear track. Follow-up work extended this to better incorporate temporal information via CNNs (Fig. 1b), with conv-PI-VAE having even higher performance54.
Behavioural analysis for neuroscience
Lightly adapting Lord Kelvin’s dictum, one may quip that ‘what you cannot measure, you cannot understand’. Consider a natural scene where various species engage in their daily activities (Fig. 3a). With our advanced primate sensory and cognitive systems, we can effortlessly extract rich semantic information from this environment: identifying the different species, characterizing the sounds they produce, interpreting their behaviour, and even detecting nuanced social dynamics such as the attentive gaze of a mother monitoring her young. As we outline below, current behavioural analysis systems are comparatively limited.
In natural scenes where various species engage in their daily activities, current analysis systems are comparatively limited in transforming animals’ behaviour into rich, structured data streams that enable straightforward enquiry through simple human-interpretable queries. a, Problem setting and solutions: localization, pose, action understanding, re-identification148 and scene-level annotations. b, Hierarchical decomposition of behaviour in a mouse, spanning three levels: activities, actions and motion primitives. At the highest level, the mouse performs three activities: self-care, Freudensprung (joy jump) and social interaction. Each activity comprises multiple actions — self-care, for instance, involves grooming and sitting upright. These actions further break down into elementary motion primitives that constitute the building blocks of movement. Drawing in a by Julia Kuhl.
Behaviour is inherently hierarchical, comprising nested sub-routines61,62,63,64, and often is not clearly discrete but, rather, continuous in nature (Fig. 3b). For example, the behaviour of a mouse colony’s social dynamics is characterized by many part-to-whole relationships both across space (from the entire colony, to individual family units, to specific mice, to their whiskers and forelimbs) and time (from seasonal reproductive cycles, to brief courtship interactions, to momentary investigative sniffs).
Ultimately, we believe that behavioural analysis systems should aim to capture this comprehensive and continuous, behavioural landscape (Fig. 3b). We advocate that the goal is to transform animals and their environment (or ecosystem) into rich, structured data streams that enable straightforward enquiry through simple human-interpretable queries. Just as a video game designer has perfect knowledge of what virtual agents perceive and how they respond to their environment, we should strive for similar insight into animal behaviour in experimental contexts.
Many of the variables we seek to measure can be inferred well from cameras (sometimes other modalities are more appropriate, but the deep learning methods work similarly) (see the section ‘Towards hybrid objectives and multimodal modelling’ for discussion of multimodality). One of the foundational (machine learning) tasks is animal detection (localization). This can be done by training detectors65,66,67, which infer bounding boxes around each individual or simple vision transformations. The latter approaches work well when the contrast is high68,69. One can also jointly estimate the location of multiple body parts, rather than just infer the body’s centre or the bounding boxes. Such pose estimation algorithms distil the geometric configuration of the animal’s body into a few user-defined keypoints70. With these methods, the locations of other objects or individuals can be inferred, thus enabling the study of how animals interact with their environment. Pose estimation is mature, widely used tools are openly available71,72,73,74,75,76 and users can improve the performance of their tailored networks by adapting the augmentation pipeline70, using post-processing or using specialized methods for crowded scenes77 (reviewed elsewhere2,7,70).
Although these tailored, specialist models extract pose within user-defined contexts, recent unified models provide keypoint spaces that work robustly across species and settings with strong zero-shot performance75, or serve as stronger initializations than standard transfer learning71 when training is necessary. Similarly, for animal detection, MegaDetector78 or Segment Anything66,67 excel at localizing and segmenting animals across videos without annotation.
Moving beyond 2D estimation, users may want to exact kinematically accurate estimates with three dimensions and even merge this with biomechanical modelling. 3D pose estimation is (typically) achieved through multiple calibrated cameras72,79,80,81,82,83, depth cameras84,85,86 or a single camera87,88,89. From a single camera one applies lifting methods, either directly from 2D pose sequences87,88,89 or with end-to-end trainable pipelines that combine multiple steps, but can even achieve excellent results for complex cases such as hand–object interactions90,91. We discuss new avenues for merging 3D pose and biomechanics (see the section ‘Towards hybrid objectives and multimodal modelling’).
After 2D or 3D pose extraction and tracking across time, activities, actions and motion primitives (Fig. 3b) — behaviours — are identified using three approaches: rule-based, supervised and unsupervised. Rule-based analysis defines behaviours through measurements — for instance, tracking head versus body keypoints enables defining heading angle and ‘look right’ behaviours, whereas tracking two mice allows defining ‘following’ heuristics. This simple yet powerful approach is widely implemented (for example, Live Mouse Tracker)92. Large language models can help researchers to write such rule-based analysis code93 (Box 2).
For supervised behavioural analysis, annotated examples of behaviour are obtained and then a classifier is trained. This classifier can operate on pose, video frames or many other modalities94,95,96. Owing to the widely available pose estimation tools, various approaches have been developed to predict behaviour from pose tracking data73,97,98,99,100,101,102. More generally, in computer science, the related task of action recognition has seen a lot of progress, due to large-scale benchmarks103 and advances in model architectures, including foundation models104,105,106,107,108 (Box 2).
For unsupervised methods, various computational approaches are widely used to decompose behaviour into ‘syllables’84,85,109,110,111,112. However, these models typically operate on a single timescale, which can be either an implicit or explicit parameter85. In unsupervised representation learning competitions for behavioural analysis, such as MABe22 (ref. 99), adapted variants of BERT113, Perceiver34, TS2Vec114 and PointNet115 initially reached the best results. In addition, AmadeusGPT93 performed well in generating rule-based analysis code from natural language user input via language models. Hierarchical masked autoencoding-based methods (hBehaveMAE)116 and contrastive methods integrating multiple timescales, such as bootstrap across multiple scales117, later reached better performance both for identifying social actions and genotype and environmental conditions.
Of course, it is (relatively) straightforward to collect a large amount of videos of animals in experiments. However, annotating these data is time consuming, costly, requires a lot of knowledge, is error prone and is subject to biases10,11,118. To develop better methods, larger data sets that annotate behaviours of interest need to be created. Here, one could also leverage published work, where the behaviour was annotated manually. Another important direction that demonstrates the power of emerging approaches is the creation of synthetic data based on simulators116,119. For example, due to the scarcity of large-scale hierarchical behavioural benchmarks, Stoffl et al.116 created a synthetic basketball playing benchmark (Shot7M2) and could show that hBehaveMAE learns interpretable behavioural latents on Shot7M2 as well as non-synthetic data sets.
Why infer all these variables when many — especially high-level behavioural inferences — are perhaps subjective and difficult to validate? Neural data offer one of the most objective metrics for assessing these measurements. The critical question is whether one can identify corresponding neural signatures in the brain. Do these signatures map hierarchically onto the circuits that generate behaviour in a hierarchical manner?
This capability would be transformative for neuroscience, where linking neural activity to naturalistic, hierarchical behaviour remains a central challenge. By providing a comprehensive behavioural read-out across multiple timescales and organizational levels, such systems would enable neuroscientists to correlate brain activity with precise behavioural events, states and decisions — dramatically advancing our understanding of neural coding, sensorimotor integration and the neural bases of behaviour. Future multimodal brain–behaviour models could tackle this.
Towards hybrid objectives and multimodal modelling
We propose a taxonomy of supervised, generative and contrastive models that can operate on neural or behavioural data alone, or jointly across modalities. Although these categories provide a useful scaffold, modern machine learning increasingly combines elements from multiple paradigms, incorporates pretrained features and trains on heterogeneous data sets (for example, CEBRA with DINO embeddings). This shift reflects a broader trend in AI: moving beyond narrowly defined tasks towards models that learn shared latent representations across diverse data streams and tasks. In neuroscience, this raises the question of whether joint brain–behaviour models might evolve along similar lines to recent successes in multimodal AI, such as vision-language models (Box 2).
In parallel to advances in neuroscience for neural and behavioural analysis, recent advances in AI, particularly in vision-language modelling, have shown the power of learning joint latent representations across modalities without assigning one as primary and others as auxiliary. Notable examples are so-called vision-language models, which are (so far) primarily used outside neuroscience. Bai et al.120 proposed an early vision-language model that combined BLIP121 (which jointly optimizes three objectives: image–text contrastive for aligning image and text embeddings; image–text matching for determining whether a caption matches an image; and language modelling for generating captions or answers from visual input) with the Qwen large language model122 (which processes visual tokens as input to the language model). Such models learn shared latent spaces by aligning visual and language streams through contrastive or generative pretraining123,124,125. These architectures capture rich semantic relationships by simultaneously encoding and decoding across modalities, offering a compelling blueprint for future neuroscience models.
In addition, the use of new AI tools for behavioural measurement has expanded rapidly in recent years. As we aimed to highlight, moving to hierarchical measurements of behaviour, and even mapping pose to biomechanical models, is now possible89,126,127,128. Namely, given a biomechanical model, one can imitate recorded 3D pose estimation data and infer muscle dynamics via physics simulations129. Naturally, inferring those (latent) variables is crucial for modelling the somatosensory, proprioceptive and motor systems, and several recent studies are at the interface of motion capture, biomechanics and neuroscience89,126,127,128. Thus, these higher-dimensional behaviour variables will be critical to reveal biological insights with joint modelling approaches.
Inspired by this, we believe that the next-generation hybrid objective models in neural data should move beyond the conventional encoder–decoder pipeline or single-modality supervision. Rather than treating spikes as outputs and behaviour as labels, or vice versa, truly multimodal neural models can learn embeddings that simultaneously predict, align and reconstruct multiple streams: spiking activity, behavioural videos or other task-related stimuli. This likely requires objective functions that integrate self-supervised contrastive, generative and reconstruction-based losses, enabling models to reason jointly about neural dynamics, internal states and externally observable behaviours. Specially, future approaches may incorporate latent dynamics with high-dimensional output modelling, where the goal is to reconstruct visual stimuli or even the biomechanical level of behaviour given neural recordings, or vice versa. Such tasks will benefit from architectural innovations beyond transformers or state-space models (Fig. 1). Although those generic architectures scale efficiently to long sequences, it is still active research in machine learning to tailor such multimodal networks to input–output multiple tasks with high performance. Also, new architectures tailored to spatio-temporal structure in neuroscience data might need to be considered. These hybrid frameworks may lead to foundation models (Box 2) that infer shared latent spaces of perception and action, enabling generalization across tasks, individuals and experimental settings.
Here, we also briefly link to data-driven and task-driven models of the brain. Work in this field also leverages the power of AI, but to explicitly build models of brain function for hypothesis testing and making discoveries (reviewed previously3,26,130). For example, recently Wang et al.131 developed a data-driven foundation model for the primary visual cortex of mice that is trained to predict spiking activity in multiple areas of the brain from measured behaviour, such as a video stimulus (animal-viewed) and pupil direction and diameter. They showed that this model generalizes to predict the response to classic visual stimuli (which was not possible before), and the responses in other mice. Notably, this model demonstrates the ability to predict cell types and anatomical areas131, illustrating the potential for multimodal applications.
Trustworthy, interpretable and performant joint models
As joint models become more central to neuroscientific discovery, we argue that it is no longer sufficient to benchmark solely on performance in spike prediction or behavioural decoding. Instead, we must systematically assess mechanistic interpretability metrics, such as ‘consistency’, ‘identifiability’ and ‘robustness’ of the models — core properties that reflect whether models yield reproducible, interpretable representations across runs, data sets and participants (Box 1). These criteria are essential for building trustworthy and scientifically useful models. Thus, future benchmarking efforts should also focus on trustworthiness and interpretability in joint brain–behaviour models, and we propose a scorecard to help shape these efforts (Table 1).
Trustworthiness derives from consistency, identifiability and robustness. Consistency across runs measures the stability of embeddings or predictions when models are retrained with different random seeds or data subsets, ensuring reproducibility54,132. Identifiability evaluates whether latent representations can be uniquely recovered up to simple transformations (for example, linear mappings) across sessions or individuals, crucial for meaningful cross-data set comparisons51,54. Robustness to noise and perturbations quantifies sensitivity to input corruption, missing data or adversarial attacks, highlighting model reliability under real-world conditions133 (Table 1). Although this is often not considered in neuroscience research, in real-world neurotechnology applications such as BMIs there is growing recognition of such issues.
Interpretability considers whether the learned features are both human-interpretable and mathematically explainable — whether attribution methods such as Shapley values or saliency maps provide consistent and faithful explanations of model decisions that generalize across data sets134,135. Moreover, recent work to expand explainable AI methods with theoretical guarantees in the time domain are emerging55,136. In addition, how well learned latent spaces correspond across different modalities (such as neural activity and behaviour) — cross-modal alignment — can be assessed137,138,139. Evaluating models in these additional dimensions could greatly aid in both tool selection for researchers, and for pushing the field to develop more interpretable models.
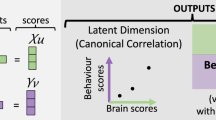
A related line of interpretability work are methods and metrics that have been developed to compare representations (Table 1). Classical methods for comparing neural population dynamics include canonical correlation analysis, which identifies linear projections that maximize shared variance between data sets140, and representational similarity analysis, which compares pairwise dissimilarity matrices of neural responses141. Centred kernel alignment was later introduced into machine learning to robustly compare representational spaces, even across layers of deep networks, and has since been shown to be mathematically related to representational similarity analysis under certain conditions142,143. Emerging methods include shape metrics, which is a very promising approach proposed by Barbosa et al.144 and Williams et al.145. In brief, this approach builds on, and formalizes, Procrustes distances146 to quantify similarity in neural populations by evaluating explicit geometric transformations between neural trajectories, allowing flexible specification of distance measures that capture population-level neural dynamics. Another metric is dynamic similarity analysis, which is a non-linear metric that compares the spatio-temporal elements of dynamical systems147.
Open challenges
Modelling across diverse neural and behavioural data types is not without complexity. As implicitly noted in this Review, challenges arise from differences in sampling rates, modality-specific noise characteristics, and methods to both assess performance and the resulting representational geometries of the models. A major challenge is the heterogeneity of data types, including spike trains, functional MRI signals and video-based pose estimation, each having different sampling rates, noise profiles, assumptions and generative mechanisms. Developing robust frameworks that can handle asynchronous, incomplete and noisy multimodal data streams remains a critical challenge.
As experimental paradigms become more naturalistic, the number of relevant behavioural measurements and variability (might) grow substantially. This creates a fundamental tension: more realistic behaviours require more complex models, but limited data necessitates simpler approaches to avoid overfitting. Cross-session modelling can help here. However, although we can now train powerful models across multiple sessions, they rely on strong assumptions. How can this be done correctly when inputs and computations vary across trials, sessions or behavioural contexts?
Model selection also remains an open problem; particularly, when ground truth latent states are unavailable, it becomes challenging to know whether the learned latents are meaningful. To aid in this, we argue that traditional metrics such as reconstruction error or decoding accuracy must be supplemented with measures such as explainability, robustness and representational similarity (Table 1). Model selection could also involve leveraging activity recorded in other brain areas. Indeed, inferring putative unmeasured inputs such as sensory inputs, neuromodulatory signals or those from upstream brain areas is a major open challenge.
Notably, interpretability is a critical challenge. Deep learning models, particularly large-scale transformers and multimodal foundation models, may not produce human-interpretable latents. As these models grow in complexity, their outputs risk becoming disconnected from mechanistic insight unless constrained by priors or structured inductive biases grounded in neuroscience.
Conclusions
In summary, we synthesized recent advances in joint modelling of neural and behavioural data, with a focus on methodological innovations, scientific and engineering motivations, and key areas for future innovation. Specifically, we discussed innovations in discriminative, generative and contrastive joint models and recent advances in behavioural analysis methods, including pose estimation and hierarchical behaviour analysis. In addition, we argued that traditional metrics such as the reconstruction error or decoding accuracy must be supplemented with measures such as explainability, robustness and representational similarity. We believe that their incorporation will yield new hybrid approaches that can leverage the rich diversity of behaviour, but also allow for new principles of neural coding to be uncovered.
Joint brain–behaviour modelling is rapidly reshaping the ability to understand how neural dynamics generate complex behaviour. Looking ahead, the fusion of discriminative, generative and contrastive approaches, large-scale neural recordings and multimodal behavioural measurements from high-level behavioural states to biomechanics promises not just better prediction but conceptual breakthroughs. Moving beyond joint models that capture the latents of neural dynamics as shaped by behaviour, future models may begin to uncover new mathematical principles of neural computation. Can emergent laws that describe how dynamic neural systems encode, transform and act on information be discovered?
The most exciting frontier lies in discovering emergent laws that describe how dynamic neural systems encode, transform and act on information — principles that might be as fundamental to neuroscience as conservation laws are to physics. As computational power grows and data become richer across ecological contexts and diverse species, we anticipate that the next generation of embodied, situated and hierarchical models will not merely simulate brain function but also reveal the organizing principles that make adaptive intelligence possible. By embracing the full complexity of natural behaviour while grounding our models in the physical reality of bodies moving through environments, we believe the field stands at the threshold of a new synthesis — one that will transform both our understanding of biological intelligence and our ability to create artificial systems that exhibit truly adaptive, flexible behaviour. The challenge ahead is not just technical but conceptual: can we develop theoretical frameworks powerful enough to bridge the gap between the richness of natural behaviour and the elegance of fundamental principles? We are optimistic that the answer is yes, and we look forward to contributing to this transformative journey.
References
Krakauer, J. W., Ghazanfar, A. A., Gomez-Marin, A., MacIver, M. A. & Poeppel, D. Neuroscience needs behavior: correcting a reductionist bias. Neuron 93, 480–490 (2017).
Pereira, T. D., Shaevitz, J. W. & Murthy, M. Quantifying behavior to understand the brain. Nat. Neurosci. 23, 1537–1549 (2020).
Mathis, M. W., Rotondo, A. P., Chang, E. F., Tolias, A. S. & Mathis, A. Decoding the brain: from neural representations to mechanistic models. Cell 187, 5814–5832 (2024).
Siegle, J. H. et al. Open Ephys: an open-source, plugin-based platform for multichannel electrophysiology. J. Neural Eng. 14, 045003 (2017).
Siegle, J. H. et al. Survey of spiking in the mouse visual system reveals functional hierarchy. Nature 592, 86–92 (2021).
Helmchen, F. & Denk, W. Deep tissue two-photon microscopy. Nat. Methods 2, 932–940 (2005).
Mathis, M. W. & Mathis, A. Deep learning tools for the measurement of animal behavior in neuroscience. Curr. Opin. Neurobiol. 60, 1–11 (2020).
Hong, G. & Lieber, C. M. Novel electrode technologies for neural recordings. Nat. Rev. Neurosci. 20, 330–345 (2019).
Manley, J. et al. Simultaneous, cortex-wide dynamics of up to 1 million neurons reveal unbounded scaling of dimensionality with neuron number. Neuron 112, 1694–1709.e5 (2024).
Anderson, D. J. & Perona, P. Toward a science of computational ethology. Neuron 84, 18–31 (2014).
von Ziegler, L., Sturman, O. & Bohacek, J. Big behavior: challenges and opportunities in a new era of deep behavior profiling. Neuropsychopharmacology 46, 33–44 (2021).
Tuia, D. et al. Perspectives in machine learning for wildlife conservation. Nat. Commun. 13, 1–15 (2022).
Couzin, I. D. & Heins, C. Emerging technologies for behavioral research in changing environments. Trends Ecol. Evol. 38, 346–354 (2023).
Jumper, J. et al. Highly accurate protein structure prediction with alphafold. Nature 596, 583–589 (2021).
Andrychowicz, M. et al. Deep learning for day forecasts from sparse observations. Preprint at https://arxiv.org/abs/2306.06079 (2023).
Vyas, S., Golub, M. D., Sussillo, D. & Shenoy, K. V. Computation through neural population dynamics. Annu. Rev. Neurosci. 43, 249–275 (2020).
Hurwitz, C. L., Kudryashova, N. N., Onken, A. & Hennig, M. H. Building population models for large-scale neural recordings: opportunities and pitfalls. Curr. Opin. Neurobiol. 70, 64–73 (2021).
Wöhr, M. & Schwarting, R. K. Affective communication in rodents: ultrasonic vocalizations as a tool for research on emotion and motivation. Cell Tissue Res. 354, 81–97 (2013).
Ishiyama, S. & Brecht, M. Neural correlates of ticklishness in the rat somatosensory cortex. Science 354, 757–760 (2016).
Hassabis, D., Kumaran, D., Summerfield, C. & Botvinick, M. Neuroscience-inspired artificial intelligence. Neuron 95, 245–258 (2017).
Mathis, M. W. Adaptive intelligence: leveraging insights from adaptive behavior in animals to build flexible AI systems. Preprint at https://arxiv.org/abs/2411.15234 (2025).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT Press, 2016).
Prince, S. J. D. Understanding Deep Learning (MIT Press, 2023).
Augustine, M. T. A survey on universal approximation theorems. Preprint at https://doi.org/10.48550/arXiv.2407.12895 (2024).
Yamins, D. L. & DiCarlo, J. J. Using goal-driven deep learning models to understand sensory cortex. Nat. Neurosci. 19, 356–365 (2016).
Schölkopf, B. & Smola, A. J. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond (MIT Press, 2002).
Ye, J. & Pandarinath, C. Representation learning for neural population activity with neural data transformers. Neurons Behav. Data Anal. Theory 5, 1–18 (2021).
Azabou, M. et al. A unified, scalable framework for neural population decoding. In Proc. 37th Conf. Neural Inf. Process. Syst. 44937–44956 (Curran Associates, 2023).
Pei, F. et al. Neural latents benchmark ’21: evaluating latent variable models of neural population activity. Preprint at https://arxiv.org/abs/2109.04463 (2022).
Zhou, Z. et al. in Human Brain Artificial Intelligence (eds Liu, Q. at al.) 192–206 (Springer, 2024).
Candelori, B. et al. Spatio-temporal transformers for decoding neural movement control. J. Neural Eng. 22, 016023 (2025).
Metzger, S. L. et al. A high-performance neuroprosthesis for speech decoding and avatar control. Nature 620, 1037–1046 (2023).
Jaegle, A. et al. Perceiver: general perception with iterative attention. In Proc. 38th Int. Conf. Mach. Learn. (eds Meila, M. & Zhang, T.) 4651–4664 (PMLR, 2021).
Vaswani, A. et al. Attention is all you need. In Proc. 31st Int. Conf. Neural Inf. Process. Syst. (eds von Luxburg, U. et al.) 6000–6010 (Curran Associates, 2017).
Havrilla, A. & Liao, W. Understanding scaling laws with statistical and approximation theory for transformer neural networks on intrinsically low-dimensional data. In Proc. 38th Conf. Neural Inf. Process. Syst. (eds Globerson, A. et al.) 42162–42210 (Curran Associates, 2024).
Shaeri, M. et al. A 2.46-mm2 miniaturized brain–machine interface (MiBMI) enabling 31-class brain-to-text decoding. IEEE J. Solid-State Circuits 59, 3566–3579 (2024).
Sani, O. G., Pesaran, B. & Shanechi, M. Dissociative and prioritized modeling of behaviorally relevant neural dynamics using recurrent neural networks. Nat. Neurosci. 27, 2033–2045 (2024).
Kingma, D. P. & Welling, M. Auto-encoding variational bayes. Preprint at https://doi.org/10.48550/arXiv.1312.6114 (2022).
Rezende, D. J., Mohamed, S. & Wierstra, D. Stochastic backpropagation and approximate inference in deep generative models. In Proc. 31st Int. Conf. Mach. Learn. (eds Xing, E. P. & Jebara, T.) 1278–1286 (PMLR, 2014).
Pandarinath, C. et al. Inferring single-trial neural population dynamics using sequential auto-encoders. Nat. Methods 15, 805–815 (2018).
Keshtkaran, M. R. et al. A large-scale neural network training framework for generalized estimation of single-trial population dynamics. Nat. Methods 19, 1572–1577 (2022).
Kato, S. et al. Global brain dynamics embed the motor command sequence of caenorhabditis elegans. Cell 163, 656–669 (2015).
Gao, Y., Archer, E., Paninski, L. & Cunningham, J. P. Linear dynamical neural population models through nonlinear embeddings. In Proc. 30th Conf. Neural Inf. Process. Syst. (Curran Associates, 2016).
Hu, A. et al. Modeling latent neural dynamics with gaussian process switching linear dynamical systems. In Proc. 38th Conf. Neural Inf. Process. Syst. (eds Globerson, A. et al.) 33805–33835 (Curran Associates, 2024).
Liu, M., Nair, A., Coria, N., Linderman, S. W. & Anderson, D. J. Encoding of female mating dynamics by a hypothalamic line attractor. Nature 634, 901–909 (2024).
Bird, A., Williams, C. K. & Hawthorne, C. Multi-task dynamical systems. J. Mach. Learn. Res. 23, 1–52 (2022).
Liu, X. et al. Self-supervised learning: generative or contrastive. IEEE Trans. Knowl. Data Eng. 35, 857–876 (2021).
Dhariwal, P. & Nichol, A. Diffusion models beat gans on image synthesis. In Proc. 35th Int. Conf. Neural Inf. Process. Syst. (eds Ranzato, M. et al.) 8780–8794 (Curran Associates, 2021).
Kapoor, J. et al. Latent diffusion for neural spiking data. In Proc. 38th Conf. Neural Inf. Process. Syst. (eds Globerson, A. et al.) 118119–118154 (Curran Associates, 2024).
Hyvärinen, A. & Pajunen, P. Nonlinear independent component analysis: Existence and uniqueness results. Neural Netw. 12, 429–439 (1999).
Oord, A. V. d., Li, Y. & Vinyals, O. Representation learning with contrastive predictive coding. Preprint at https://doi.org/10.48550/arXiv.1807.03748 (2019).
Zimmermann, R. S., Sharma, Y., Schneider, S., Bethge, M. & Brendel, W. Contrastive learning inverts the data generating process. In Proc. 38th Int. Conf. Mach. Learn. (eds Meila, M. & Zhang, T.) 12979–12990 (PMLR, 2021).
Schneider, S., Lee, J. H. & Mathis, M. W. Learnable latent embeddings for joint behavioural and neural analysis. Nature 617, 360–368 (2023).
Schneider, S., Laiz, R. G., Filippova, A., Frey, M. & Mathis, M. W. Time-series attribution maps with regularized contrastive learning. In Proc. 28th Int. Conf. Artif. Intell. Stat. (PMLR, 2025).
Chen, T., Kornblith, S., Norouzi, M. & Hinton, G. A simple framework for contrastive learning of visual representations. In Proc. 37th Int. Conf. Mach. Learn. (eds Daumé III, H. & Singh, A.) 1597–1607 (PMLR, 2020).
Laiz, R. G., Schmidt, T. & Schneider, S. Self-supervised contrastive learning performs non-linear system identification. In Proc. 13th Int. Conf. Learn. Represent. (ICLR, 2025).
Gosztolai, A., Peach, R. L., Arnaudon, A., Barahona, M. & Vandergheynst, P. Marble: interpretable representations of neural population dynamics using geometric deep learning. Nat. Methods 22, 612–620 (2025).
Zhou, D. & Wei, X. Learning identifiable and interpretable latent models of high-dimensional neural activity using PI-VAE. In Proc. 35th Int. Conf. Neural Inf. Process. Syst. (Curran Associates, 2020).
Khemakhem, I., Kingma, D. P. & Hyvärinen, A. Variational autoencoders and nonlinear ICA: a unifying framework. In Proc. Int. Conf. Artif. Intell. Stat. (PMLR, 2019).
Lashley, K. S. et al. The Problem of Serial Order in Behavior Vol. 21 (Bobbs-Merrill, 1951).
Tinbergen, N. On aims and methods of ethology. Z. Tierpsychol. 20, 410–433 (1963).
Botvinick, M. M. Hierarchical models of behavior and prefrontal function. Trends Cogn. Sci. 12, 201–208 (2008).
Winter, D.Biomechanics and Motor Control of Human Movement (Wiley, 2009).
Girshick, R., Donahue, J., Darrell, T. & Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proc. IEEE Conf. Comput. Vision Pattern Recogn. 580–587 (IEEE, 2014).
Kirillov, A. et al. Segment anything. In Proc. IEEE Conf. Comput. Vision 4015–4026 (IEEE, 2023).
Ravi, N. et al. SAM 2: segment anything in images and videos. In Proc. 13th Int. Conf. Learn. Represent. (ICLR, 2025).
Romero-Ferrero, F., Bergomi, M. G., Hinz, R. C., Heras, F. J. & de Polavieja, G. G. idtracker. ai: tracking all individuals in small or large collectives of unmarked animals. Nat. Methods 16, 179 (2019).
Walter, T. & Couzin, I. D. TRex, a fast multi-animal tracking system with markerless identification, and 2D estimation of posture and visual fields. eLife 10, e64000 (2021).
Mathis, A., Schneider, S., Lauer, J. & Mathis, M. W. A primer on motion capture with deep learning: principles, pitfalls, and perspectives. Neuron 108, 44–65 (2020).
Mathis, A. et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281 (2018).
Nath, T. et al. Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat. Protoc. 14, 2152–2176 (2019).
Segalin, C. et al. The Mouse Action Recognition System (MARS) software pipeline for automated analysis of social behaviors in mice. eLife 10, e63720 (2021).
Lauer, J. et al. Multi-animal pose estimation, identification and tracking with deeplabcut. Nat. Methods 19, 496–504 (2022).
Ye, S. et al. Superanimal pretrained pose estimation models for behavioral analysis. Nat. Commun. 15, 5165 (2024).
Pereira, T. D. et al. SLEAP: a deep learning system for multi-animal pose tracking. Nat. Methods 19, 486–495 (2022).
Zhou, M., Stoffl, L., Mathis, M. W. & Mathis, A. Rethinking pose estimation in crowds: overcoming the detection information bottleneck and ambiguity. In Proc. IEEE Conf. Comput. Vision 14689–14699 (IEEE, 2023).
Beery, S., Morris, D. & Yang, S. Efficient pipeline for camera trap image review. Preprint at https://doi.org/10.48550/arXiv.1907.06772 (2019).
Dunn, T. W. et al. Geometric deep learning enables 3D kinematic profiling across species and environments. Nat. Methods 18, 564–573 (2021).
Karashchuk, P. et al. Anipose: a toolkit for robust markerless 3D pose estimation. Cell Rep. 36, 109730 (2021).
Joska, D. et al. AcinoSet: a 3D pose estimation dataset and baseline models for cheetahs in the wild. In 2021 IEEE Int. Conf. Robot. Autom. (ICRA) 13901–13908 (IEEE, 2021).
Kaneko, T. et al. Deciphering social traits and pathophysiological conditions from natural behaviors in common marmosets. Curr. Biol. 34, 2854–2867 (2024).
Yurimoto, T. et al. Development of a 3D tracking system for multiple marmosets under free-moving conditions. Commun. Biol. 7, 216 (2024).
Wiltschko, A. B. et al. Mapping sub-second structure in mouse behavior. Neuron 88, 1121–1135 (2015).
Weinreb, C. et al. Keypoint-MoSeq: parsing behavior by linking point tracking to pose dynamics. Nat. Methods 21, 1329–1339 (2024).
Menegas, W. et al. High-throughput unsupervised quantification of patterns in the natural behavior of marmosets. eLife 13, RP103586 (2024).
Gosztolai, A. et al. LiftPose3D, a deep learning-based approach for transforming two-dimensional to three-dimensional poses in laboratory animals. Nat. Methods 18, 975–981 (2021).
Hu, B. et al. 3D mouse pose from single-view video and a new dataset. Sci. Rep. 13, 13554 (2023).
DeWolf, T., Schneider, S., Soubiran, P., Roggenbach, A. & Mathis, M. W. Neuro-musculoskeletal modeling reveals muscle-level neural dynamics of adaptive learning in sensorimotor cortex. Preprint at bioRxiv https://doi.org/10.1101/2024.09.11.612513 (2024).
Hampali, S., Sarkar, S. D., Rad, M. & Lepetit, V. Keypoint transformer: solving joint identification in challenging hands and object interactions for accurate 3D pose estimation. In Proc. IEEE/CVF Conf. Comput. Vision Pattern Recogn. (CVPR) 11090–11100 (IEEE, 2022).
Qi, H., Zhao, C., Salzmann, M. & Mathis, A. HOISDF: constraining 3D hand–object pose estimation with global signed distance fields. In Proc. IEEE/CVF Conf. Comput. Vision Pattern Recogn. (CVPR) 10392–10402 (IEEE, 2024).
de Chaumont, F. et al. Real-time analysis of the behaviour of groups of mice via a depth-sensing camera and machine learning. Nat. Biomed. Eng. 3, 930–942 (2019).
Ye, S., Lauer, J., Zhou, M., Mathis, A. & Mathis, M. AmadeusGPT: a natural language interface for interactive animal behavioral analysis. In Proc. 37th Int. Conf. Neural Inf. Process. Syst. 6297-6329 (Curran Associates, 2023).
Ding, G., Sener, F. & Yao, A. Temporal action segmentation: an analysis of modern techniques. IEEE Trans. Pattern Anal Mach. Intell. 46, 1011–1030 (2023).
Bohnslav, J. P. et al. DeepEthogram, a machine learning pipeline for supervised behavior classification from raw pixels. eLife 10, e63377 (2021).
Camilleri, M. P., Bains, R. S. & Williams, C. K. Of mice and mates: automated classification and modelling of mouse behaviour in groups using a single model across cages. Int. J. Comput. Vis. 132, 5491–5513 (2024).
Kabra, M., Robie, A. A., Rivera-Alba, M., Branson, S. & Branson, K. JAABA: interactive machine learning for automatic annotation of animal behavior. Nat. Methods 10, 64 (2013).
Sturman, O. et al. Deep learning-based behavioral analysis reaches human accuracy and is capable of outperforming commercial solutions. Neuropsychopharmacology 45, 1942–1952 (2020).
Sun, J. J. et al. MABe22: a multi-species multi-task benchmark for learned representations of behavior. In Proc. 40th Int. Conf. Mach. Learn. (eds Krause, A. et al.) 32936–32990 (PMLR, 2023).
Bordes, J. et al. Automatically annotated motion tracking identifies a distinct social behavioral profile following chronic social defeat stress. Nat. Commun. 14, 4319 (2023).
Goodwin, N. L. et al. Simple behavioral analysis (SimBA) as a platform for explainable machine learning in behavioral neuroscience. Nat. Neurosci. 27, 1411–1424 (2024).
Kozlova, E., Bonnetto, A. & Mathis, A. DLC2Action: a deep learning-based toolbox for automated behavior segmentation. Preprint at bioRxiv https://doi.org/10.1101/2025.09.27.678941 (2025).
Madan, N., Moegelmose, A., Modi, R., Rawat, Y. S. & Moeslund, T. B. Foundation models for video understanding: a survey. Preprint at arXiv https://doi.org/10.48550/arXiv.2405.03770 (2024).
Feichtenhofer, C., Fan, H., Malik, J. & He, K. Slowfast networks for video recognition. In Proc. EEE/CVF Conf. Comput. Vision Pattern Recogn. (CVPR) 6202–6211 (2019).
Zhu, W. et al. MotionBERT: a unified perspective on learning human motion representations. In Proc. EEE/CVF Conf. Comput. Vision Pattern Recogn. (CVPR) 15085–15099 (2023).
He, K. et al. Masked autoencoders are scalable vision learners. In Proc. EEE/CVF Conf. Comput. Vision Pattern Recogn. (CVPR) 16000–16009 (2022).
Feichtenhofer, C. et al. Masked autoencoders as spatiotemporal learners. Adv. Neural Inf. Process. Syst. 35, 35946–35958 (2022).
Tong, Z., Song, Y., Wang, J. & Wang, L. VideoMAE: masked autoencoders are data-efficient learners for self-supervised video pre-training. In Proc. 36th Int. Conf. Neural Inf. Process. Syst. (Curran Associates, 2022).
Berman, G. J., Choi, D. M., Bialek, W. & Shaevitz, J. W. Mapping the stereotyped behaviour of freely moving fruit flies. J. R. Soc. Interface. 11, 20140672 (2014).
Markowitz, J. E. et al. The striatum organizes 3D behavior via moment-to-moment action selection. Cell 174, 44–58 (2018).
Hsu, A. I. & Yttri, E. A. B-SOiD, an open-source unsupervised algorithm for identification and fast prediction of behaviors. Nat. Commun. 12, 5188 (2021).
Luxem, K. et al. Identifying behavioral structure from deep variational embeddings of animal motion. Commun. Biol. 5, 1267 (2022).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conf. North Am. Chapter Assoc. Comput. Linguist. 4171–4186 (ACL, 2019).
Yue, Z. et al. TS2Vec: towards universal representation of time series. In Proc. AAAI Conf. Artif. Intel. 8980–8987 (2022).
Qi, C. R., Su, H., Mo, K. & Guibas, L. J. PointNet: deep learning on point sets for 3D classification and segmentation. In Proc. IEEE Conf. Comput. Vision Pattern Recogn. 652–660 (2017).
Stoffl, L., Bonnetto, A., d’Ascoli, S. & Mathis, A. Elucidating the hierarchical nature of behavior with masked autoencoders. In European Conf. Comput. Vision 106–125 (Springer, 2024).
Azabou, M. et al. Relax, it doesn’t matter how you get there: a new self-supervised approach for multi-timescale behavior analysis. In Proc. 37th Int. Conf. Neural Inf. Process. Syst. 28491–28509 (Curran Associates, 2023).
Tuyttens, F. et al. Observer bias in animal behaviour research: can we believe what we score, if we score what we believe? Anim. Behav. 90, 273–280 (2014).
De Melo, C. M. et al. Next-generation deep learning based on simulators and synthetic data. Trends Cogn. Sci. 26, 174–187 (2022).
Bai, J. et al. Qwen-VL: a versatile vision-language model for understanding, localization, text reading, and beyond. Preprint at https://arxiv.org/abs/2308.12966 (2023).
Li, J., Li, D., Xiong, C. & Hoi, S. Blip: Bootstrapping language-image pre-training for unified vision-language understanding and generation. In Proc. Int. Conf. Mach. Learn. 12888–12900 (PMLR, 2022).
Bai, J. et al. Qwen technical report. Preprint at https://arxiv.org/abs/2309.16609 (2023).
Li, F. et al. LLaVA-NeXT-Interleave: tackling multi-image, video, and 3D in large multimodal models. Preprint at arXiv https://doi.org/10.48550/arXiv.2407.07895 (2024).
Li, B. et al. LLaVA-OneVision: easy visual task transfer. Preprint at https://arxiv.org/abs/2408.03326 (2024).
Ye, S., Qi, H., Mathis, A. & Mathis, M. W. LLaVAction: evaluating and training multi-modal large language models for action recognition. Preprint at https://arxiv.org/abs/2503.18712 (2025).
Vargas, A. M. et al. Task-driven neural network models predict neural dynamics of proprioception. Cell 187, 1745–1761 (2024).
Melis, J. M., Siwanowicz, I. & Dickinson, M. H. Machine learning reveals the control mechanics of an insect wing hinge. Nature 628, 795–803 (2024).
Vaxenburg, R. et al. Whole-body physics simulation of fruit fly locomotion. Nature 643, 1312–1320 (2025).
Buchanan, T. S., Lloyd, D. G., Manal, K. & Besier, T. F. Neuromusculoskeletal modeling: estimation of muscle forces and joint moments and movements from measurements of neural command. J. Appl. Biomech. 20, 367–395 (2004).
Doerig, A. et al. The neuroconnectionist research programme. Nat. Rev. Neurosci. 24, 431–450 (2023).
Wang, E. Y. et al. Foundation model of neural activity predicts response to new stimulus types. Nature 640, 470–477 (2025).
Lipton, Z. C. The mythos of model interpretability: in machine learning, the concept of interpretability is both important and slippery. Queue 16, 31–57 (2018).
Goodfellow, I. J., Shlens, J. & Szegedy, C. Explaining and harnessing adversarial examples. Preprint at arXiv https://doi.org/10.48550/arXiv.1412.6572 (2014).
Ribeiro, M. T., Singh, S. & Guestrin, C. “Why should I trust you?” Explaining the predictions of any classifier. In Proc. 22nd ACM SIGKDD Int. Conf. Knowl. Discov. Data Mining 1135–1144 (ACM, 2016).
Lundberg, S. M. & Lee, S.-I. A unified approach to interpreting model predictions. In Proc. 31st Int. Conf. Neural Inf. Process. Syst. (Curran Associates, 2017).
Jang, H., Kim, C. & Yang, E. Timing: Temporality-aware integrated gradients for time series explanation. In ICLR 2025 Workshop XAI4Sci. (ICLR, 2025).
Jazayeri, M. & Ostojic, S. Interpreting neural computations by examining intrinsic and embedding dimensionality of neural activity. Curr. Opin. Neurobiol. 70, 113–120 (2021).
Abid, A., Zhang, M. J., Bagaria, V. K. & Zou, J. Y. Exploring patterns enriched in a dataset with contrastive principal component analysis. Nat. Commun. 9, 2134 (2018).
Merk, T. et al. Invasive neurophysiology and whole brain connectomics for neural decoding in patients with brain implants. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-025-01467-9 (2025).
Raghu, M., Gilmer, J., Yosinski, J. & Sohl-Dickstein, J. SVCCA: singular vector canonical correlation analysis for deep learning dynamics and interpretability. In Proc. 31st Int. Conf. Neural Inf. Process. Syst. (Curran Associates, 2017).
Kriegeskorte, N., Mur, M. & Bandettini, P. A. Representational similarity analysis — connecting the branches of systems neuroscience. Front. Syst. Neurosci. https://doi.org/10.3389/neuro.06.004.2008 (2008).
Kornblith, S., Norouzi, M., Lee, H. & Hinton, G. Similarity of neural network representations revisited. In Proc. Int. Conf. Mach. Learn. 3519–3529 (PMLR, 2019).
Williams, A. H. Equivalence between representational similarity analysis, centered kernel alignment, and canonical correlations analysis. In Proc. UniReps 2nd Edn Workshop Unif. Represent. Neural Models (PMLR, 2024).
Barbosa, J. et al. Quantifying differences in neural population activity with shape metrics. Preprint at bioRxiv https://doi.org/10.1101/2025.01.10.632411 (2025).
Williams, A. H., Kunz, E. M., Kornblith, S. & Linderman, S. W. Generalized shape metrics on neural representations. Adv. Neural Inf. Process. Syst. 34, 4738–4750 (2021).
Schönemann, P. H. A generalized solution of the orthogonal procrustes problem. Psychometrika 31, 1–10 (1966).
Ostrow, M., Eisen, A., Kozachkov, L. & Fiete, I. Beyond geometry: comparing the temporal structure of computation in neural circuits with dynamical similarity analysis. In Proc. 37th Int. Conf. Neural Inf. Process. Syst. 33824–33837 (Curran Associates, 2023).
Vidal, M., Wolf, N., Rosenberg, B., Harris, B. P. & Mathis, A. Perspectives on individual animal identification from biology and computer vision. Integr. Comp. Biol. 61, 900–916 (2021).
Locatello, F., Bauer, S., Lucic, M., Raetsch, G., Gelly, S., Schölkopf, B. & Bachem, O. Challenging common assumptions in the unsupervised learning of disentangled representations. In Proc. Int. Conf. Mach. Learn. 4114–4124 (PMLR, 2019).
Radford, A. & Narasimhan, K. Improving language understanding by generative pre-training. Preprint at https://arxiv.org/abs/1801.06146 (2018).
OpenAI et al. GPT-4 technical report. Preprint at https://doi.org/10.48550/arXiv.2303.08774 (2024).
Radford, A. et al. Learning transferable visual models from natural language supervision. In Proc. Int. Conf. Mach. Learn. 8748–8763 (PMLR, 2021).
Hyvärinen, A., Sasaki, H. & Turner, R. E. Nonlinear ICA using auxiliary variables and generalized contrastive learning. In 22nd Int. Conf. Artif. Intell. Stat. 859–868 (PMLR, 2019).
Brown, T. et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 33, 1877–1901 (2020).
Bommasani, R. et al. On the opportunities and risks of foundation models. Preprint at https://arxiv.org/abs/2108.07258 (2021).
Castro, P. S. et al. Discovering symbolic cognitive models from human and animal behavior. Preprint at bioRxiv https://doi.org/10.1101/2025.02.05.636732 (2025).
Acknowledgements
The authors thank members of their laboratories, especially M. Simos, P. Muratore and H. Mirzaeri for discussions. This work was funded by the Swiss National Science Foundation (SNSF) though grants 310030_212516 (to A.M.), TMSGI3_226525 (to M.W.M.) and 320030-227871 (to A.M. and M.W.M.).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article. A.M. led the behavioural modelling sections, and M.W.M. led the neural and joint modelling sections.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Neuroscience thanks Michael Yartsev, who co-reviewed with Adam Lowet, and the other, anonymous, reviewer for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
European Charter for the Responsible Development of Neurotechnologies: https://www.braincouncil.eu/european-charter-for-the-responsible-development-of-neurotechnologies/
Glossary
- Agent-based systems
-
Artificial intelligence (AI) systems capable of autonomous goal-directed behaviour, including planning, reasoning and interaction with their environment to achieve specified objectives.
- Attribution methods
-
Techniques that identify which input features contribute most to a model’s output.
- Decoder
-
A network module that transforms latent representations back into the data domain, reconstructing or generating outputs.
- Deep learning
-
A subset of machine learning using multilayer neural networks to learn complex, hierarchical data representations.
- Digital twin
-
A computational replica of a real system used for simulation, prediction or control.
- Discrete state transitions
-
Changes between distinct system states, often modelled as jumps in state-space dynamics.
- Embeddings
-
Vector representations capturing semantic or structural relationships among data elements.
- Encoder
-
A network module that maps inputs into a latent space.
- Latent space
-
The abstract representation space where encoded data are organized by learned features. Latent representations live in the latent space, just as integers live in the set of integers \({\mathbb{Z}}\). It is a space, because it also has structure. For instance, often you can add two latent representations, or take the average.
- Machine learning
-
Algorithms that learn patterns from data to make predictions or decisions without explicit programming.
- Neural dynamics
-
The time-evolving activity patterns and interactions among neurons or artificial network units.
- Poisson loss
-
A likelihood-based loss for count data assuming Poisson-distributed observations. Commonly used to model spike counts in neuroscience.
- Poisson noise
-
Random variability in count data arising from discrete stochastic events.
- Self-supervised learning
-
Learning representations from unlabelled data such as by predicting masked parts of the input from other parts, or learning from temporal structure.
- Supervised learning
-
Learning from labelled data pairs (x, y) to map inputs x to known outputs y.
- Topological data analysis
-
Method using topology to characterize the shape and structure of complex data.
- Universal approximators
-
Given enough capacity, neural networks can approximate any continuous function on compact domains to arbitrary precision. For example, even a feedforward network with a single hidden layer of sufficient width is a universal approximator.
- Zero-shot performance
-
The performance of a model when evaluated on tasks or samples without training data (from this task/setting). This evaluates generalization. Few-shot evaluation allows training on a few samples.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mathis, M.W., Mathis, A. Joint modelling of brain and behaviour dynamics with artificial intelligence. Nat. Rev. Neurosci. 27, 87–100 (2026). https://doi.org/10.1038/s41583-025-00996-1
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41583-025-00996-1
This article is cited by
-
Leveraging insights from neuroscience to build adaptive artificial intelligence
Nature Neuroscience (2026)