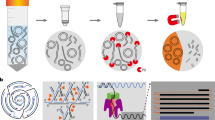
Abstract
Extrachromosomal circular DNA elements (eccDNAs) have been described in the literature for several decades, and are known for their broad existence across different species1,2. However, their biogenesis and functions are largely unknown. By developing a new circular DNA enrichment method, here we purified and sequenced full-length eccDNAs with Nanopore sequencing. We found that eccDNAs map across the entire genome in a close to random manner, suggesting a biogenesis mechanism of random ligation of genomic DNA fragments. Consistent with this idea, we found that apoptosis inducers can increase eccDNA generation, which is dependent on apoptotic DNA fragmentation followed by ligation by DNA ligase 3. Importantly, we demonstrated that eccDNAs can function as potent innate immunostimulants in a manner that is independent of eccDNA sequence but dependent on eccDNA circularity and the cytosolic DNA sensor Sting. Collectively, our study not only revealed the origin, biogenesis and immunostimulant function of eccDNAs but also uncovered their sensing pathway and potential clinical implications in immune response.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The eccDNA sequencing data and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) with accession number GSE165919.
Code availability
The code used to analyse eccDNA sequencing data is available at GitHub (https://github.com/YiZhang-lab/eccDNA_RCA_nanopore).
References
Paulsen, T., Kumar, P., Koseoglu, M. M. & Dutta, A. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 34, 270–278 (2018).
Gaubatz, J. W. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat. Res. 237, 271–292 (1990).
Hotta, Y. & Bassel, A. Molecular size and circularity of DNA in cells of mammals and higher plants. Proc. Natl Acad. Sci. USA 53, 356–362 (1965).
Moller, H. D. et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun. 9, 1069 (2018).
Dillon, L. W. et al. Production of extrachromosomal microDNAs is linked to mismatch repair pathways and transcriptional activity. Cell Rep. 11, 1749–1759 (2015).
Hull, R. M. et al. Transcription-induced formation of extrachromosomal DNA during yeast ageing. PLoS Biol. 17, e3000471 (2019).
Gresham, D. et al. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc. Natl Acad. Sci. USA 107, 18551–18556 (2010).
Sinclair, D. A. & Guarente, L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91, 1033–1042, (1997).
Shibata, Y. et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 336, 82–86 (2012).
Zhu, J. et al. Molecular characterization of cell-free eccDNAs in human plasma. Sci. Rep. 7, 10968 (2017).
Cloninger, C., Felton, M., Paul, B., Hirakawa, Y. & Metzenberg, S. Control of pH during plasmid preparation by alkaline lysis of Escherichia coli. Anal. Biochem. 378, 224–225 (2008).
DeLap, R. J., Rush, M. G., Zouzias, D. & Khan, S. Isolation and preliminary characterization of the small circular DNA present in African green monkey kidney (BSC-1) cells. Plasmid 1, 508–521 (1978).
Landry, J. J. et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 3, 1213–1224 (2013).
Tichy, E. D. Mechanisms maintaining genomic integrity in embryonic stem cells and induced pluripotent stem cells. Exp. Biol. Med. 236, 987–996 (2011).
Rang, F. J., Kloosterman, W. P. & de Ridder, J. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 19, 90 (2018).
Joffroy, B., Uca, Y. O., Presern, D., Doye, J. P. K. & Schmidt, T. L. Rolling circle amplification shows a sinusoidal template length-dependent amplification bias. Nucleic Acids Res. 46, 538–545 (2018).
Soh, Y. Q. et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159, 800–813 (2014).
Marichal, T. et al. DNA released from dying host cells mediates aluminum adjuvant activity. Nat. Med. 17, 996–1002 (2011).
Sakahira, H., Enari, M. & Nagata, S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391, 96–99 (1998).
Li, L. Y., Luo, X. & Wang, X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412, 95–99 (2001).
Shiokawa, D., Kobayashi, T. & Tanuma, S. Involvement of DNase γ in apoptosis associated with myogenic differentiation of C2C12 cells. J. Biol. Chem. 277, 31031–31037 (2002).
Arakawa, H. et al. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic Acids Res. 40, 2599–2610, (2012).
Lu, G. et al. Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proc. Natl Acad. Sci. USA 113, 1256–1260 (2016).
Simsek, D. et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 471, 245–248 (2011).
Li, Y., Berke, I. C. & Modis, Y. DNA binding to proteolytically activated TLR9 is sequence-independent and enhanced by DNA curvature. EMBO J. 31, 919–931 (2012).
Lange, S. S. & Vasquez, K. M. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Mol. Carcinog. 48, 571–580 (2009).
Wang, Z. et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl Acad. Sci. USA 105, 5477–5482 (2008).
Green, D. R., Ferguson, T., Zitvogel, L. & Kroemer, G. Immunogenic and tolerogenic cell death. Nat. Rev. Immunol. 9, 353–363 (2009).
Sundaresan, R., Parameshwaran, H. P., Yogesha, S. D., Keilbarth, M. W. & Rajan, R. RNA-independent DNA cleavage activities of Cas9 and Cas12a. Cell Rep. 21, 3728–3739 (2017).
Putney, S. D., Benkovic, S. J. & Schimmel, P. R. A DNA fragment with an α-phosphorothioate nucleotide at one end is asymmetrically blocked from digestion by exonuclease III and can be replicated in vivo. Proc. Natl Acad. Sci. USA 78, 7350–7354 (1981).
Sauer, J. D. et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694 (2011).
Hou, B., Reizis, B. & DeFranco, A. L. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity 29, 272–282 (2008).
Oberhammer, F. et al. Apoptotic death in epithelial cells: cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 12, 3679–3684 (1993).
Shiokawa, D. & Tanuma, S. Differential DNases are selectively used in neuronal apoptosis depending on the differentiation state. Cell Death Differ. 11, 1112–1120 (2004).
Wang, H. et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 65, 4020–4030 (2005).
Cox, D., Yuncken, C. & Spriggs, A. I. Minute chromatin bodies in malignant tumours of childhood. Lancet 1, 55–58 (1965).
Turner, K. M. et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125 (2017).
Li, X. D. et al. Pivotal roles of cGAS–cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 (2013).
Andreeva, L. et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein–DNA ladders. Nature 549, 394–398 (2017).
Tovey, M. G., Lallemand, C. & Thyphronitis, G. Adjuvant activity of type I interferons. Biol. Chem. 389, 541–545 (2008).
Moore, J. B. & June, C. H. Cytokine release syndrome in severe COVID-19. Science 368, 473–474 (2020).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ignatiadis, N., Klaus, B., Zaugg, J. B. & Huber, W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13, 577–580 (2016).
Du, Q., Kotlyar, A. & Vologodskii, A. Kinking the double helix by bending deformation. Nucleic Acids Res. 36, 1120–1128 (2008).
Liu, D. et al. Branched kissing loops for the construction of diverse RNA homooligomeric nanostructures. Nat. Chem. 12, 249–259 (2020).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Gel, B. & Serra, E. karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33, 3088–3090 (2017).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Acknowledgements
We thank D. Schatz (Yale University) for the antibody to mouse Lig4, R. Rajan (University of Oklahoma) for the initial gift of FnoCas protein, X. Zhang for help in BMDC preparation, P. Yin for allowing us to use the AFM instrument and J. Lieberman for critical reading of the manuscript. H.C. was supported by a Leukemia and Lymphoma Society Fellow Award. D.L. is a Merck Fellow of the Life Sciences Research Foundation. This work was supported by the Howard Hughes Medical Institute. F.W.A. and Y.Z. are Investigators of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
Y.Z. conceived the project; Y.W. and Y.Z. designed the experiments; Y.W. performed the majority of the experiments; H.C. generated the ligase-knockout cell lines; M.W. developed the method to call eccDNA from Nanopore data and performed all bioinformatics analyses; M.N.D. performed pilot analysis of eccDNA sequencing data; D.L. helped with the SAFM imaging; F.W.A. supervised the work by H.C.; Y.W. and Y.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
A patent application (with the patent agency Wolf, Greenfield & Sacks) covering the eccDNA purification method and the application of eccDNA as an immunostimulatant has been filed by the Boston Children’s Medical Center Corporation.
Additional information
Peer review information Nature thanks Birgitte Regenberg and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Selective binding of eccDNAs to beads and high sensitivity of DNA detection by SYBR Gold staining.
a, Representative gel image showing selective binding of circular DNA to magnetic silica beads in solution A. The assay was tested with 200× (mass) linear DNA (lane 4 and 6) and 1X (mass) DNA circles (lane 5 and 6). Lanes 1-3 indicate the input DNAs. Lane 1, 0.5% of input linear DNA for lane 4 was loaded; lane 2, equal amount of input circular DNAs for lane 5 were loaded; lane 3, 0.5% of input linear DNA and equal amount of input circular DNAs for lane 6 were loaded. Lanes 4-6 were DNAs recovered from Solution A. lane 4, 200× (mass) linear DNA alone undergone purification by solution A; lane 5, 1× (mass) DNA circles undergone purification by solution A; lane 6 mixture of 200× (mass) linear DNA and 1× (mass) DNA circles undergone purification by solution A. Note, only circular DNAs were recovered. The recovery rate was very high, particularly for the smaller circular DNAs, C2: a 200 bp circle, C5: a 525 bp circle. b, High sensitivity of DNA detection using vertical agarose gel electrophoresis and SYBR Gold staining. 0.2 μl commercial DNA ladder (total of 10 ng) was undergone 5× series dilutions and fractionated by 1% vertical agarose gel electrophoresis, stained by SYBR Gold, DNA in lane-4 were 125× diluted of that in lane-1, and could still be visualized. The detection limit is estimated as 5-10 pg/band. Results in a–b are representative of three independent experiments.
Extended Data Fig. 2 Summary of the nanopore sequencing data.
a, Diagram of nanopore long read sequencing of eccDNA. Tandem copies of eccDNAs were self-concatenated to long molecule by rolling cycle amplification (RCA), and directly read through by Oxford Nanopore. Each copy of eccDNA molecule in a single long molecule was sequenced multiple times. “S”: split site. b, Summary of mESC eccDNA long reads from an Oxford Nanopore MinION flow cell. c, Diagram showing how the eccDNA full-length sequence is called and categorized. Nanopore reads from (a), the dashed box, were aligned to a single locus (Continuous) or to multiple loci (Non-continuous) in genome. Continuous: an example of six sub-regions of a long nanopore read repeatedly aligned to a single locus, where full length sequence of eccDNA was presented with only one split site (S); Non-continuous: an example of eight sub-regions of a single long read sequentially aligned to two separate loci (Locus-1 and Locus-2), representing an eccDNA ligated by two genomic fragments (2f ecc) with two split sites (S1 and S2). d, Criteria for eccDNA calling. EccDNAs were called based on their number of full passes aligned to the genome. Long reads with less than two full passes were discarded (left and middle panel). Left, Nanopore read that hits genome only once either fails to designate the genomic start and end site of eccDNA (up panel), or miss the middle region (bottom panel) that may or may not include in the original eccDNA molecule (Uncertain); Middle, because of potential sequencing error of Oxford Nanopore, reads that hit genome more than once but less than twice (1 full pass) were also discarded due to the lack of confirmation in eccDNA calling, particularly on designating the start and end site of eccDNA. Right, eccDNA molecules were called from long reads when covered at least twice (>=2 full pass) on their aligned loci.
Extended Data Fig. 3 Summary of Illumina short read sequencing and Chromosomal distribution of eccDNA fragments.
a, Summary of eccDNA short read sequencing without RCA. Purified eccDNA was directly tagmented with Tn5 (Illumina Nextera), and sequenced with Illumina Hiseq 2500 in PE150 mode. b, Chromosomal distribution of eccDNA short reads.
Extended Data Fig. 4 Effects of Endonuclease G, DNase γ, and DNA ligases knockout on apoptosis and eccDNA generation.
a, Quantification of eccDNA from cells of the indicated treatment. EccDNA production was presented as nanogram per 10 million cells; ud, under detection limit. Bars indicate mean ± s.e.m. of three independent experiments. b, Diagram illustration and PCR confirmation of the EndoG knockout mESC line. Gene structure of EndoG, sgRNAs (red arrows), two sets of screen primers for internal sites (black arrowhead, “n” =negative knockout cell line) and external sites (red arrowhead, “p” =positive knockout cell line) were shown. c, Diagram illustration and PCR confirmation of the DNase γ knockout mESC lines. Gene structure of DNase γ, sgRNAs (red arrows), two sets of screen primers for internal sites (black arrowhead, “n” =negative knockout cell line) and external sites (red arrowhead, “p” =positive knockout cell line) were shown. d, Cell viability is not affected by EndoG or DNase γ KO. Cell viability was evaluated by flow cytometry, after staining with Far Red Live/Dead Cell Stain Kit, FSC-A/SSC-A and FSC-A/FSC-H were sequentially used to exclude debris and gate on singlets, respectively, then gate on APC+ as dead cells. Bars indicate mean ± s.e.m. of dead cell ratio from three independent experiments. e, Deficiency of DNase γ or EndoG in mESC do not significantly alter UV-induced cell death. Cell death was measured by flow cytometry, after staining with Far Red Live/Dead Cell Stain Kit, FSC-A/SSC-A and FSC-A/FSC-H were sequentially used to exclude debris and gate on singlets, respectively, then gate on APC+ as dead cells. Bar indicate mean ± s.e.m. of dead cell ratio of 3 independent experiments. f, Quantification of eccDNAs presented in Fig. 3d. EccDNAs below the mtDNA band were quantified by densitometry (by Image J 1.53e) and presented as relative levels to that in WT cells of parallel eccDNA purifications. Bars indicate mean ± s.e.m. of three independent experiments. g, Quantification of eccDNA below the mtDNA band by densitometry (by Image J 1.53e). Data are presented as relative levels to that purified from WT cells of parallel eccDNA purifications (Fig. 3g), bars indicate mean ± s.e.m. of three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparison test was used for multiple comparisons in e, f, g. **, p<0.01; ns, no significant.
Extended Data Fig. 5 EccDNAs are potent immunostimulants for BMDCs.
a, Confirmation of BMDC identity by flow cytometry. After differentiating bone marrow cells with 20 ng/ml GM-CSF for 7 days, cell debris (left) were first excluded and singlets (middle) were then phenotyped on the basis of their CD11c and major histocompatibility complex II (MHCII) expression (right) to define BMDC, the numbers indicate the percentage of gated cells. Data were further analyzed with FlowJo. V10.8.0 and shown as representative of 2 independent experiments. b, Various DNAs were resolved by agarose gel electrophoresis. Li-DNA: sheared linear genomic DNA; poly(dG:dC), poly(deoxyguanylic-deoxycytidylic). Representative gel of three independent experiments was shown. c, d, Bar graphs showing the relative mRNA (c) and protein (ELISA) (d) levels of IL-6 and TNF-α. Data is shown as mean ± s.e.m. of replicates (n=4 per group) of a representative experiment in three independent experiments. e, f, DNase I pretreatment abolishes IL-6 and TNF-α induction by eccDNA. Equal amount of DNA (120 ng/ml) used to prepare transfection mixtures were pre-treated with or without DNase I as indicated, then transfected to BMDCs. 12 h later, total RNA was extracted for RT-qPCR (e) and medium was collected for ELISA (f). Data are shown as mean ± s.e.m. of replicates (n=4 per group) of a representative experiment in three independent experiments. Comparisons in c–f were done on biological replicates by Ordinary one-way ANOVA with Tukey’s multiple comparison test. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, no significant; nd, not detected.
Extended Data Fig. 6 EccDNAs are potent immunostimulants for BMDMs.
a, Confirmation of BMDMs identify by flow cytometry. After differentiating bone marrow cells with L929 conditioned medium for 7 days, cells debris (left) were first excluded and singlets (middle) were phenotyped on the basis of F4/80 and CD11b expression (right) to define BMDM, the numbers indicate the percentage of gated cells. Data were further analyzed with FlowJo. V10.8.0 and shown as representative of 2 independent experiments. b, c, EccDNAs induce cytokine genes in BMDMs. Bar graphs showing the induction of mRNA (b) and protein (c) of IL-6 and TNF-α in BMDMs that transfected with varying levels of eccDNA, compared with fragmented linear DNA, and poly(dG:dC). RT-qPCR were performed after 12-hours transfection. ELISA analysis (c) was performed after 24-hours transfection. Data are shown as mean ± s.e.m. of replicates (n=4 per group) of a representative experiment of three independent experiments. Comparisons were performed on biological replicates with equal amount of DNA transfection by Ordinary one-way ANOVA with Tukey’s multiple comparison test. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, no significant; nd, not detected.
Extended Data Fig. 7 The circularity of eccDNAs, but not the sequence, is critical for their immunostimulant activity.
a, Bar graphs showing the relative IL-6 and TNF-α mRNA levels. Data is shown as mean ± s.e.m. of replicates (n=4 per group) of a representative experiment in three independent experiments. b, c, Bar graphs showing the relative IL-6 and TNF-α mRNA levels (b), and protein (ELISA) levels (c). Data is shown as mean ± s.e.m. of replicates (n=4 per group) of a representative experiment in three independent experiments. d, Diagram of DNA transfection efficiency and stability assay. Step-1: 5 Phosphorothioate (purple “*”) end-protected synthetic linear DNA (PS-syn-linear) and its circular form with equal number of phosphorothioate bonds (PS-Syn-circular) were transfected to BMDCs in the same way as in Fig. 5 for either 1 or 12 h. Step-2: cells were lysed in 100 μl lysis buffer and treated with Thermoliable Proteinase K to prepare total DNA. Step-3: 4 μl total DNA was used for qPCR with a set of primers that amplify a 117 bp fragment in both linear and circular DNA. e, qPCR analysis of the samples prepared as described in (d). Transfected DNA level was normalized to that of 10 ng/ml PS-syn-linear transfection (n= 4 or 6, as indicated by dotes). Bars indicate mean ± s.e.m. of three independent experiments. f, 12 h after transfection of indicated DNA at 30 ng/ml, medium was collected for ELISA essay, Data are shown as mean ± s.e.m. of replicates (n=4 per group) of a representative experiment in three independent experiments. Comparisons in a–c were performed on biological replicates by Ordinary one-way ANOVA with Tukey’s multiple comparison test; two-tailed unpaired t tests (e, f). *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, no significant.
Extended Data Fig. 8 Reproducibility of the BMDC transcriptomes.
a–c, Scatter plot of pair-wise comparison of the transcriptomes of three independent BMDC samples of untreated (a), treated with eccDNA (b) or linear DNA (c). The x and y-axis are log2(FPKM+0.1).
Extended Data Fig. 9 Global transcriptional response to eccDNA in BMDMs.
a–c, Scatter plot of pair-wise comparison of the transcriptomes of three independent BMDM samples of untreated (a), treated with eccDNA (b) or linear DNA (Extended Data Fig. 6). The x and y-axis are log2(FPKM+0.1). d, Scatter plot showing 380 genes (left panel, red dots) that are significantly induced (FC ≥ 5, adjusted p-value < 0.001; FC and p-value were generated using DESeq242 and p-values were adjusted by IHW43) by eccDNA, but not linear DNA, in BMDMs. 24 significantly induced cytokine genes are indicated (right panel, red dots). The x and y-axis are log2-transformed normalized read counts. e, Heatmap presentation of the top 20 induced genes. f, GO terms enriched in the genes activated by eccDNA treatment in BMDMs. The number of genes in each term and the p-value of the enrichment is indicated.
Extended Data Fig. 10 Analysis of the eccDNA sensing pathway.
a, Scatter plot indicates the transcriptomes of the two replicates of synthetic circular DNA treated BMDCs are highly similar. The x and y-axis are log2(FPKM+0.1). b, Western blot confirmation of the Sting-/- and Myd88-/- cells. Blots were performed once, but cell genotypes were further confirmed by mRNA sequences from RNA-seq data (data not shown). c, Correlation of the transcriptomes of 2 replicates eccDNA treated Sting-/- and Myd88-/- BMDC. The x and y-axis are log2(FPKM+0.1).
Supplementary information
Supplementary Fig. 1
This file contains uncropped images of agarose gels and scan images of immunoblotting.
Supplementary Table 1
List of CRISPR–Cas9 guide RNA and genotyping PCR primer sequences for the different knockout cell lines
Supplementary Table 2
List of RT–qPCR primer sequences for the various cytokine genes analysed
Supplementary Table 3
List of synthetic small DNAs and their PCR amplification primer sequences
Supplementary Table 4
List of the differentially expressed genes in eccDNA-treated BMDCs
Supplementary Table 5
List of the differentially expressed genes in eccDNA-treated BMDMs
Supplementary Table 6
List of the differentially expressed genes in eccDNA-treated BMDCs in response to Sting1 or Myd88 knockout
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Wang, M., Djekidel, M.N. et al. eccDNAs are apoptotic products with high innate immunostimulatory activity. Nature 599, 308–314 (2021). https://doi.org/10.1038/s41586-021-04009-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-021-04009-w
This article is cited by
-
Detection of primary cancer types via fragment size selection in circulating cell-free extrachromosomal circular DNA
Genome Medicine (2026)
-
Mitotic errors as triggers of cell death and inflammation
Nature Cell Biology (2026)
-
Compartment-specific eccDNA patterns reveal senescence associated biomarkers in hUC-MSCs
Scientific Reports (2026)
-
Opportunities and challenges of targeting cGAS–STING in cancer
Nature Reviews Cancer (2026)
-
Machine learning identified extrachromosomal DNA-related 12 gene signatures to predict cancer immunotherapy response
Cancer Cell International (2025)