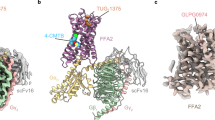
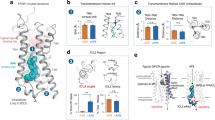
Abstract
Free fatty acid receptor 2 (FFA2) is a G protein-coupled receptor (GPCR) that is a primary sensor for short-chain fatty acids produced by gut microbiota. Consequently, FFA2 is a promising drug target for immunometabolic disorders1,2,3,4. Here we report cryogenic electronic microscopy structures of FFA2 in complex with two G proteins and three distinct classes of positive allosteric modulators (PAMs), and describe noncanonical activation mechanisms that involve conserved structural features of class A GPCRs. Two PAMs disrupt the E/DRY activation microswitch5 and stabilize the conformation of intracellular loop 2 by binding to lipid-facing pockets near the cytoplasmic side of the receptor. By contrast, the third PAM promotes the separation of transmembrane helices 6 and 7 by interacting with transmembrane helix 6 at the receptor–lipid interface. Molecular dynamic simulations and mutagenesis experiments confirm these noncanonical activation mechanisms. Furthermore, we demonstrate the molecular basis for the Gi versus Gq bias, which is due to distinct conformations of intracellular loop 2 stabilized by different PAMs. These findings provide a framework for the design of tailored GPCR modulators, with implications that extend beyond FFA2 to the broader field of GPCR drug discovery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The 3D cryo-EM density maps of the structures of FFA2 signalling complexes have been deposited into the Electron Microscopy Data Bank under the accession numbers EMD-45732 for TUG-1375–4-CMTB–FFA2–miniGq, EMD-45738 for TUG-1375–compound 187–FFA2–miniGq, EMD-45743 for TUG-1375–AZ-1729–FFA2–Gi and EMD-49745 for TUG-1375–compound 187–FFA2–Gi. Atomic coordinates for the atomic models have been deposited into the PDB under the accession numbers 9CLW for TUG-1375–4-CMTB–FFA2–miniGq, 9CM3 for TUG-1375–compound 187–FFA2–miniGq, 9CM7 for TUG-1375–AZ-1729–FFA2–Gi and 9NS9 for TUG-1375–compound 187–FFA2–Gi. The following structural data used for comparisons are available from the PDB: 7LD3 (the A1R bound to the PAM MIPS521)38; 8J22 (TUG-1375-bound FFA2 coupled with Gi)22; 8J23 (empty FFA2 coupled with Gi); 8T3S (C4-bound FFA2 coupled with miniGq)21; 8J21 (C4-bound FFA3 coupled with Gi)22; 8G05 (GPR84 coupled with Gi)86; and 6C1R (C5aR bound to PMX53 and avacopan)60. Simulation trajectories were uploaded to GPCRmd (https://www.gpcrmd.org/)87 and linked to the current paper. Simulation identifiers are provided in Supplementary Table 3.
References
Stoddart, L. A., Smith, N. J. & Milligan, G. International Union of Pharmacology. LXXI. Free fatty acid receptors FFA1, -2, and -3: pharmacology and pathophysiological functions. Pharmacol. Rev. 60, 405–417 (2008).
Alvarez-Curto, E. & Milligan, G. Metabolism meets immunity: the role of free fatty acid receptors in the immune system. Biochem. Pharmacol. 114, 3–13 (2016).
Kimura, I., Ichimura, A., Ohue-Kitano, R. & Igarashi, M. Free fatty acid receptors in health and disease. Physiol. Rev. 100, 171–210 (2020).
McCarville, J. L., Chen, G. Y., Cuevas, V. D., Troha, K. & Ayres, J. S. Microbiota metabolites in health and disease. Annu. Rev. Immunol. 38, 147–170 (2020).
Zhou, Q. et al. Common activation mechanism of class A GPCRs. eLife 8, e50279 (2019).
Milligan, G., Shimpukade, B., Ulven, T. & Hudson, B. D. Complex pharmacology of free fatty acid receptors. Chem. Rev. 117, 67–110 (2017).
Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009).
Kimura, I. et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 4, 1829 (2013).
Zhao, Y. et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 11, 752–762 (2018).
Antunes, K. H. et al. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun. 10, 3273 (2019).
Kimura, I., Inoue, D., Hirano, K. & Tsujimoto, G. The SCFA receptor GPR43 and energy metabolism. Front. Endocrinol. 5, 85 (2014).
Bindels, L. B., Dewulf, E. M. & Delzenne, N. M. GPR43/FFA2: physiopathological relevance and therapeutic prospects. Trends Pharmacol. Sci. 34, 226–232 (2013).
Huang, S. et al. Lithium carbonate alleviates colon inflammation through modulating gut microbiota and Treg cells in a GPR43-dependent manner. Pharmacol. Res. 175, 105992 (2022).
Park, B. O. et al. Novel GPR43 agonists exert an anti-Inflammatory effect in a colitis model. Biomol. Ther. 30, 48–54 (2022).
Hansen, A. H. et al. Discovery of a potent thiazolidine free fatty acid receptor 2 agonist with favorable pharmacokinetic properties. J. Med. Chem. 61, 9534–9550 (2018).
Lee, T. et al. Identification and functional characterization of allosteric agonists for the G protein-coupled receptor FFA2. Mol. Pharmacol. 74, 1599–1609 (2008).
Smith, N. J. et al. Extracellular loop 2 of the free fatty acid receptor 2 mediates allosterism of a phenylacetamide ago-allosteric modulator. Mol. Pharmacol. 80, 163–173 (2011).
Milligan, G., Bolognini, D. & Sergeev, E. in Free Fatty Acid Receptors (eds Milligan, G. & Kimura, I) 17–32 (Springer, 2017).
Bolognini, D. et al. A novel allosteric activator of free fatty acid 2 receptor displays unique Gi-functional bias. J. Biol. Chem. 291, 18915–18931 (2016).
Park, B. O. et al. The short-chain fatty acid receptor GPR43 modulates YAP/TAZ via RhoA. Mol. Cells 44, 458–467 (2021).
Zhang, X. et al. Structural basis for the ligand recognition and signaling of free fatty acid receptors. Sci. Adv. 10, eadj2384 (2024).
Li, F. et al. Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3. Cell Res. 34, 323–326 (2024).
Brown, A. J. et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 278, 11312–11319 (2003).
Hudson, B. D. et al. Chemically engineering ligand selectivity at the free fatty acid receptor 2 based on pharmacological variation between species orthologs. FASEB J. 26, 4951–4965 (2012).
Wang, Y. et al. The first synthetic agonists of FFA2: discovery and SAR of phenylacetamides as allosteric modulators. Bioorg. Med. Chem. Lett. 20, 493–498 (2010).
Lind, S. et al. Interdependent allosteric free fatty acid receptor 2 modulators synergistically induce functional selective activation and desensitization in neutrophils. Biochim. Biophys. Acta Mol. Cell. Res. 1867, 118689 (2020).
Olsen, R. H. J. et al. TRUPATH, an open-source biosensor platform for interrogating the GPCR transducerome. Nat. Chem. Biol. 16, 841–849 (2020).
Sergeev, E. et al. Non-equivalence of key positively charged residues of the free fatty acid 2 receptor in the recognition and function of agonist versus antagonist ligands. J. Biol. Chem. 291, 303–317 (2016).
Kim, K. et al. Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell 182, 1574–1588 (2020).
Xia, R. et al. Cryo-EM structure of the human histamine H1 receptor/Gq complex. Nat. Commun. 12, 2086 (2021).
Meyerowitz, J. G. et al. The oxytocin signaling complex reveals a molecular switch for cation dependence. Nat. Struct. Mol. Biol. 29, 274–281 (2022).
Nehme, R. et al. Mini-G proteins: novel tools for studying GPCRs in their active conformation. PLoS ONE 12, e0175642 (2017).
Wan, Q. et al. Mini G protein probes for active G protein-coupled receptors (GPCRs) in live cells. J. Biol. Chem. 293, 7466–7473 (2018).
Duan, J. et al. Cryo-EM structure of an activated VIP1 receptor–G protein complex revealed by a NanoBiT tethering strategy. Nat. Commun. 11, 4121 (2020).
Koehl, A. et al. Structure of the µ-opioid receptor–Gi protein complex. Nature 558, 547–552 (2018).
Swaminath, G. et al. Mutational analysis of G-protein coupled receptor—FFA2. Biochem. Biophys. Res. Commun. 405, 122–127 (2011).
Grundmann, M. et al. A molecular mechanism for sequential activation of a G protein-coupled receptor. Cell Chem. Biol. 23, 392–403 (2016).
Draper-Joyce, C. J. et al. Positive allosteric mechanisms of adenosine A1 receptor-mediated analgesia. Nature 597, 571–576 (2021).
Ballesteros, J. A. & Weinstein, H. in Methods in Neurosciences Vol. 25 (ed. Sealfon, S. C.) 366–428 (Academic Press, 1995).
Wang, Y., Yu, Z., Xiao, W., Lu, S. & Zhang, J. Allosteric binding sites at the receptor–lipid bilayer interface: novel targets for GPCR drug discovery. Drug Discov. Today 26, 690–703 (2021).
Ding, T., Karlov, D. S., Pino-Angeles, A. & Tikhonova, I. G. Intermolecular interactions in G protein-coupled receptor allosteric sites at the membrane interface from molecular dynamics simulations and quantum chemical calculations. J. Chem. Inf. Model. 62, 4736–4747 (2022).
Ciancetta, A. et al. Probe confined dynamic mapping for G protein-coupled receptor allosteric site prediction. ACS Cent. Sci. 7, 1847–1862 (2021).
Filipek, S. Molecular switches in GPCRs. Curr. Opin. Struct. Biol. 55, 114–120 (2019).
Barker, G. et al. 3-Substituted 2-amino-indole derivatives. WIPO patent WO2015/198045 (2015).
Weis, W. I. & Kobilka, B. K. The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 87, 897–919 (2018).
Hauser, A. S. et al. GPCR activation mechanisms across classes and macro/microscales. Nat. Struct. Mol. Biol. 28, 879–888 (2021).
Manglik, A. & Kruse, A. C. Structural basis for G protein-coupled receptor activation. Biochemistry 56, 5628–5634 (2017).
Lu, J. et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat. Struct. Mol. Biol. 24, 570–577 (2017).
Ho, J. D. et al. Structural basis for GPR40 allosteric agonism and incretin stimulation. Nat. Commun. 9, 1645 (2018).
Powers, A. S. et al. A non-canonical mechanism of GPCR activation. Nat. Commun. 15, 9938 (2024).
Slosky, L. M., Caron, M. G. & Barak, L. S. Biased allosteric modulators: new frontiers in GPCR drug discovery. Trends Pharmacol. Sci. 42, 283–299 (2021).
Flock, T. et al. Selectivity determinants of GPCR–G-protein binding. Nature 545, 317–322 (2017).
Inoue, A. et al. Illuminating G-protein-coupling selectivity of GPCRs. Cell 177, 1933–1947 (2019).
Flock, T. et al. Universal allosteric mechanism for Gα activation by GPCRs. Nature 524, 173–179 (2015).
Bahia, D. S. et al. Hydrophobicity of residue351 of the G protein Gi1α determines the extent of activation by the α2A-adrenoceptor. Biochemistry 37, 11555–11562 (1998).
Manglik, A. & Kobilka, B. The role of protein dynamics in GPCR function: insights from the β2AR and rhodopsin. Curr. Opin. Cell Biol. 27, 136–143 (2014).
Hilger, D. The role of structural dynamics in GPCR-mediated signaling. FEBS J. 288, 2461–2489 (2021).
Zhuang, Y. et al. Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor. Cell Res. 31, 593–596 (2021).
Liu, X. et al. Mechanism of β2AR regulation by an intracellular positive allosteric modulator. Science 364, 1283–1287 (2019).
Liu, H. et al. Orthosteric and allosteric action of the C5a receptor antagonists. Nat. Struct. Mol. Biol. 25, 472–481 (2018).
Latorraca, N. R. et al. Molecular mechanism of GPCR-mediated arrestin activation. Nature 557, 452–456 (2018).
Liu, P. et al. The structural basis of the dominant negative phenotype of the Gαi1β1γ2 G203A/A326S heterotrimer. Acta Pharmacol. Sin. 37, 1259–1272 (2016).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Stoddart, L. A., Smith, N. J., Jenkins, L., Brown, A. J. & Milligan, G. Conserved polar residues in transmembrane domains V, VI, and VII of free fatty acid receptor 2 and free fatty acid receptor 3 are required for the binding and function of short chain fatty acids. J. Biol. Chem. 283, 32913–32924 (2008).
Bolognini, D. et al. Chemogenetics defines receptor-mediated functions of short chain free fatty acids. Nat. Chem. Biol. 15, 489–498 (2019).
Glide Release 2021-3 (Schrödinger, 2021).
Maestro Release 2021-3 (Schrödinger, 2021).
AMBER 2023 (Univ. California, San Francisco, 2023).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Doerr, S., Harvey, M. J., Noe, F. & De Fabritiis, G. HTMD: high-throughput molecular dynamics for molecular discovery. J. Chem. Theory Comput. 12, 1845–1852 (2016).
Lee, J. et al. CHARMM-GUI supports the Amber force fields. J. Chem. Phys. 153, 035103 (2020).
AMBER 2020 (Univ. California, San Francisco, 2020).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Tian, C. et al. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J. Chem. Theory Comput. 16, 528–552 (2020).
Dickson, C. J., Walker, R. C. & Gould, I. R. Lipid21: complex lipid membrane simulations with AMBER. J. Chem. Theory Comput. 18, 1726–1736 (2022).
He, X., Man, V. H., Yang, W., Lee, T. S. & Wang, J. A fast and high-quality charge model for the next generation general AMBER force field. J. Chem. Phys. 153, 114502 (2020).
Gowers, R. J et al. MDAnalysis: A Python package for the rapid analysis of molecular dynamics simulations. In Proc. 15th Python in Science Conference (eds Benthall, S. & Rostrup, S.) 98–105 (SciPy, 2016).
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Zhang, X. et al. Pro-phagocytic function and structural basis of GPR84 signaling. Nat. Commun. 14, 5706 (2023).
Rodriguez-Espigares, I. et al. GPCRmd uncovers the dynamics of the 3D-GPCRome. Nat. Methods 17, 777–787 (2020).
Acknowledgements
We thank the cryo-EM facility directed by J. Conway and partially supported by the grants S10 OD025009 (Krios) and S10 OD019995 (Falcon 2/3 camera) from the National Institutes of Health (NIH). This work was supported by NIH grant R35GM128641 (to C.Z.), Medical Research Council grant MR/X010198/1 (to G.M.), Biotechnology and Biological Sciences Research Council grants BB/R001480/1 and BB/S000453/1 (to G.M.) and BB/R007101/1 (to I.G.T.), Lundbeck Foundation grant R307-2018-2950 (to E.R.U.), and the Novo Nordisk Foundation grant NNF21OC0069019 (to T.U.). A.-A.G.’s PhD study is supported by the MSCA COFUND CITI-GENS Programme funded by the EU Horizon 2020 research and innovation programme (grant agreement no. 945231). This project made use of computational time on Kelvin-2 supported by the Engineering and Physical Sciences Research Council (EPSRC) (grant numbers EP/T022175/1 and EP/W03204X/1) and ARCHER2 granted through the UK High-End Computing Consortium for Biomolecular Simulation, HECBioSim (https://www.hecbiosim.ac.uk), supported by the EPSRC (grant numbers EP/R029407/1 and EP/W03204X/1).
Author information
Authors and Affiliations
Contributions
C.Z., G.M. and I.G.T. conceived the project and designed the research with X.Z. X.Z. performed protein expression and purification studies, screened the cryo-EM grids, collected the cryo-EM data and processed the data under the supervision of C.Z. L.J. and S.M. performed pharmacological and mutagenesis studies under the supervision of G.M. A.-A.G. performed the computational analyses under the supervision of I.G.T. G.M. and I.G.T. analysed data. A.V. and K.S.-K. performed compound synthesis and pharmacological characterizations under the supervision of T.U. and E.R.U. C.Z. wrote the manuscript together with G.M. and I.G.T. with help from X.Z., A.-A.G., L.J., E.R.U. and T.U.
Corresponding authors
Ethics declarations
Competing interests
G.M. and T.U. are co-founders and directors of Caldan Therapeutics (https://www.caldantherapeutics.com/). G.M. is a co-founder and director of KelticPharmaTherapeutics (https://keltic-pharma.com/). Both companies have interests in the development of FFA4 activators. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Asuka Inoue, Maria Marti-Solano and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Pharmacological characterization of FFA2 ligands.
a. β-arrestin 1 recruitment induced by FFA2 activators and modulators. b. β-arrestin 2 recruitment induced by FFA2 activators and modulators in wild-type HEK293T and G protein deficient (ΔGs, ΔG12, ΔG13, ΔGq, ΔG11 + Pertussis toxin treatment to inactivate Gi-family G proteins) HEK293T cells. Data are means +/– S.E.M. n = 3 (three biologically independent experiments). c. Cooperativity of function between three FFA2 PAMs and the orthosteric agonist C3 (propionate). The ability of the indicated concentrations of 4-CMTB, compound 187, and AZ-1729 to modulate inhibition of forskolin-stimulated levels of cAMP via FFA2 is shown. Data are means +/– S.E.M. n = 3 (three biologically independent experiments). d. Binding characteristics of FFA2 modulators assessed in co-operativity studies. aAgonist refers to the compound used to generate concentration-response curve. bModulator is the compound used in defined concentrations. cpKA represents values estimated for the agonist. dpKB represents values estimated for the modulator. eGain in potency was calculated using the equation (pEC50 agonist + highest concentration of modulator) - (pEC50 agonist + vehicle). fGain in efficacy was calculated using the equation (Emax agonist + highest concentration of modulator) - (Emax agonist + vehicle). nd means not determined (in experiments in which the effect of compound 187 was largely manifest in terms of efficacy and thus affinity values could not be derived). Emax was constrained to the maximal possible system response and n was constrained to 1 (the slope factor). Statistical significance was determined using one-way ANOVA followed by Dunnett’s multiple comparisons test; *p < 0.05, **p < 0.01, ****p < 0.0001, ns denotes not significant. Cmpd187 refers to compound 187.
Extended Data Fig. 2 Cryo-EM data processing and analysis.
a, Representative cryo-EM micrograph (scale bar: 50 nm) and 2D class averages (scale bar: 5 nm). The micrograph shown is one example of 5535 micrographs for miniGq-coupled FFA2 bound to TUG-1375 and 4-CMTB. b-m, Cryo-EM image processing workflow for TUG-1375-4-CMTB-FFA2-miniGq complex (b-d), TUG-1375-compound 187-FFA2-miniGq complex (e-g), TUG-1375-AZ-1729-FFA2-Gi complex (h-j), and TUG-1375-compound 187-FFA2-Gi complex (k-m), respectively. Detailed information on protein purification is provided in Supplementary Fig. 1. Angular distributions of the particles used in the final reconstruction are shown in c for TUG-1375-4-CMTB-FFA2-miniGq complex; f for TUG-1375-compound 187-FFA2-miniGq complex; i for TUG-1375-AZ-1729-FFA2-Gi complex; and l for TUG-1375-compound 187-FFA2-Gi complex. Gold-standard Fourier shell correlation (FSC) curves are shown in d for TUG-1375-4-CMTB-FFA2-miniGq complex; g for TUG-1375-compound 187-FFA2-miniGq complex; j for TUG-1375-AZ-1729-FFA2-Gi complex; and m for TUG-1375-compound 187-FFA2-Gi complex. n, Cryo-EM maps and models of the seven transmembrane helices (TM1-7) of miniGq-coupled FFA2 bound to TUG-1375 and 4-CMTB. Representative cryo-EM maps of FFA2 in other complexes and ligands are shown in Supplementary Fig. 2.
Extended Data Fig. 3 TUG-1375 binding in the orthosteric binding pocket.
a, Left and middle panels show the comparison of the receptor interaction profiles of TUG-1375 and butyrate (C4) with FFA2. The binding profile of TUG-1375 is based on the structure of miniGq-coupled FFA2 in complex with TUG-1375 and compound 187, while the C4 binding profile is derived from our previously published structure (PDB ID: 8T3S). Polar interactions are shown as black dashed lines. Right panel shows the outward movement of TM4 in the structure of FFA2 with TUG-1375 compared to that with C4 (butyrate) indicated by the red arrow. The red circle indicates the group of TUG-1375 that causes such movement. B, Effects of mutations within the orthosteric pocket on the function of TUG-1375 measured by β-arrestin 2 recruitment. Detailed calculations based on the concentration-response curves (upper) are summarized in the table (lower). pEC50 of TUG-1375 and efficacy compared to that of TUG-1375 at wild type (WT) human FFA2 was assessed for the indicated point mutants of the receptor by β-arrestin 2 recruitment assays. For the indicated mutants, the ability of compound 187 to inhibit forskolin stimulated levels of cAMP in cells stably expressing eYFP-tagged forms of the receptor was used to confirm expression and function of an appropriately processed and cell surface delivered form of FFA2. Data are means +/– s.e.m. n = 3 (three biologically independent experiments). Statistical significance was assessed by one way ANOVA followed by Dunnett’s multiple comparison test. * p < 0.05, ** p < 0.01, *** p < 0.001, ****p < 0.0001. NR means no detectable response. c, MD simulations on TUG-1375 binding. The upper panel shows a representative simulation frame depicting TUG-1375 in the orthosteric binding pocket, with key interacting residues shown as sticks. Residue carbon colors and stick thickness represent average electrostatic and van der Waals interaction energies between each residue and the ligand, respectively. Dashed lines indicate hydrogen bonds. The lower panel shows violin plots indicating the distribution of distances between the guanidyl fragment of R1805.39 and the carboxylic group of E166ECL2 (left) and between the Cα atoms of Y165ECL2 and Y903.33 (right) across three 1 μs replicates of FFA2/4-CMTB/TUG-1375/miniGq (Full complex), FFA2/TUG-1375 (TUG-1375 only, without the G protein), and empty FFA2 (Empty, without the ligand and G protein). The results revealed that TUG-1375 affects the ECL2 conformation and the top of TM4. Upon ligand removal, the orthosteric pocket shrinks significantly due to a major conformational change in ECL2, with E166ECL2 and Y165ECL2 shifting towards R1805.39 and Y903.33, respectively, forming extensive electrostatic and aromatic contacts. Detailed analysis of TUG-1375 binding can be found in Supplementary Information. Cmpd187 refers to compound 187.
Extended Data Fig. 4 Binding properties of FFA2 ago-PAMs at Site 1 and 2.
a, Loss of agonist activity and cooperativity with TUG-1375 for 4-CMTB at N230D FFA2, as measured by cAMP reduction assays. The data in the top row show that 4-CMTB lacks agonist action at N230D FFA2 whilst TUG-1375, compound 187 and AZ-1729 all activate this mutant as effectively as wild type. The data in the left and the middle panel of the bottom row indicate that 4-CMTB does not produce co-operativity with TUG-1375 at N230D FFA2 (left), while such effects of both AZ-1729 and compound 187 are retained at this mutant (middle). The data in the right panel indicate that the human FFA2 antagonist [3H]GLPG0974 displays high affinity binding to both wild type and N230D FFA2. Data are means +/– S.E.M. n = 3 (three biologically independent experiments). b, Structural comparison of FFA2 and FFA3 at Site 1. FFA2 and FFA3 are shown in green and pink, respectively. The space in FFA3 (middle panel, based on the structure PDB ID 8J21) corresponding to Site 1 in FFA2 (left panel) adopts a distinct conformation, which would result in a steric clash with 4-CMTB if it were to adopt a similar binding pose (right panel). c, Co-solvent simulations of AZ-1729 and compound 187. The middle figure shows the orthosteric ligand C4 and the probes as they are placed before the simulation, along with the cylinder (semi-transparent, blue) confining the probes around the area of interest, around ICL2. The top left figure shows AZ-1729 and the probe that allowed to identify its interaction with E1063.49. The top right figure shows compound 187 and its probe molecule. The bottom left and right figures show the snapshots of the probe-AZ and probe-187, respectively, in MD simulations with the semi-transparent surface denoting the volume occupied by the probe for > 20% of the simulation time. d, Structural alignment of Site 2 for compound 187 in the structures with miniGq and Gi. The interaction profile of compound 187 and the overall conformation of Site 2 including ICL2 are almost identical in these two structures. e, Effects of mutations in Site 2 on the potency of FFA2 modulators measured by [35S]GTPγS binding assays. Data are means +/– s.e.m. for at least 3 experiments. pEC50 and Emax values are listed in Extended Data Table 2. f, Undetectable β-arrestin-2 recruitment by AZ-1729 at Site 2 mutations in FFA2. β-arrestin-2 recruitment assays were performed with varying concentrations of TUG-1375 or AZ-1729 at wild type, E106G, and G102V FFA2. Data are means +/– s.e.m. n = 3 (three biologically independent experiments). Cmpd187 refers to compound 187.
Extended Data Fig. 5 Molecular dynamics analysis of FFA2 conformational changes and interactions upon different ligand binding and G protein coupling.
a-c, Molecular interactions between FFA2 ICL2 and G protein. a, Hydrophobic contacts between ICL2 (P114-R121) and G protein α5 helix. Data show mean number of carbon-carbon contacts ( ≤ 4 Å) per frame ± s.e.m. from three independent 1 μs simulations (n = 3). Individual replicate values shown as black dots. AZ-1729/Gi complex shows enhanced contacts compared to TUG-1375 alone, with significant reduction in Y117A mutant. b, Hydrogen between ICL2 and G protein α5 helix. Data show mean number of hydrogen bonds (D-A distance ≤3.5 Å, angle 180 ± 30°) per frame ± s.e.m. from three independent 1 μs simulations (n = 3). Individual replicate values shown as black dots. AZ-1729 enhances hydrogen bonding with Gi, while R121A mutation shows the strongest reduction in these interactions. c, Hydrogen bonds between R121ICL2 and D350H5.22 of Gi. Data show mean number of hydrogen bonds per frame ± s.e.m. from three independent 1 μs simulations (n = 3). Individual replicate values shown as black dots. Enhanced interactions in AZ-1729 complex and complete loss with R121A mutation. d-e, Analysis of R3.50 interactions with Y5.58 and the distance between R3.50 and D/NP7.50xxY motif in FFA2. d, Sidechain polar contacts between R1073.50 and Y1995.58. Data show percentage of frames with nitrogen-to-oxygen distance <4.5 Å ± s.e.m. from three independent 1 μs simulations (n = 3). Individual replicate values shown as black dots. e, Proximity between R1073.50 guanidyl carbon and P2707.50 Cα. Data show percentage of frames with distance <14 Å ± s.e.m. from three independent 1 μs simulations (n = 3). Individual replicate values shown as black dots. f-k, Conformational stability analysis of ICL2. f-g, Root-mean-square deviation (RMSD) of ICL2 residues P114-R121. Data show mean RMSD ± s.e.m. from three independent 1 μs simulations (n = 3), calculated after alignment to minimized cryo-EM structures using 7-transmembrane bundle Cα atoms. Individual replicate values shown as points. h-i, Root-mean-square fluctuation (RMSF) of ICL2 residues P114-R121. Data show mean RMSF ± s.e.m. from three independent 1 μs simulations (n = 3), calculated after alignment using 7-transmembrane bundle Cα atoms. Individual replicate values shown as points. j, RMSD distributions of ICL2 (P114-R121) relative to FFA2/TUG-1375/AZ-1729/Gi1 complex. Violin plots show probability densities from 3 simulation replicates per complex. Horizontal bars indicate minimum, maximum, and median values. Percentages indicate proportion of frames with RMSD < 1.5 Å. k, Similar analysis using FFA2/TUG-1375/compound 187/miniGq complex as a reference. l-n, Analysis of polar contacts at the TM6-TM7 interface. Violin plots show distributions of inter-atomic distances from snapshots taken every 100 ps across three independent 1 μs simulations per complex. When multiple atoms are selected (e.g., Oδ1 and Nδ2 of N230), the shortest distance between residue atoms is shown. Horizontal bars indicate minimum, maximum, and median values. l, Distances between N2306.43 and N2657.45 (D/NP7.50xxY motif) with percentage of frames ≤3.5 Å indicated. m, Oxygen-to-sulfur distances between N2306.43 and C2346.47 (C6.47W6.48xP6.50 motif) with percentage of frames ≤4.5 Å indicated. n, Hydrogen bonds between cytosolic region of TM6 (Q215-N230) and TM7 (R255-Y274). Data show percentage of frames with at least one hydrogen bond present ± s.e.m. from three independent 1 μs simulations (n = 3). Individual replicate values shown as black dots. Conformational stability analysis of FFA2 receptor states can be found in Supplementary Fig. 3. Detailed RMSD and RMSF data for all simulated systems and residue-ligand interaction energy values can be found in Supplementary Tables 1 and 2. Cmpd187 refers to compound 187.
Extended Data Fig. 6 G protein-coupling to FFA2.
a, Interactions between FFA2 and the α5 helix of Gi (from the structure of FFA2 with AZ-1729 and Gi) or miniGq (from the structure of FFA2 with compound187 and miniGq). b, Interactions between FFA2 and the αN helix of Gi or miniGq. Polar interactions are shown as black dashed lines. Specifically, in both structures, D350H5.22 (G protein numbering) in Gi, or E242H5.22 in mGαi/s/q forms a polar interaction with R121ICL2 in FFA2. Additionally, L348H5.20 and L353H5.25 in Gi form hydrophobic interactions with V1113.54, F2025.61, M2065.65, and L2236.36 of FFA2, while the corresponding residues, L240H5.20 and L245H5.25 in mGαi/s/q, also form direct interactions with these residues in FFA2. Notably, residues P114ICL2, V115ICL2, and L119ICL2 in ICL2 of FFA2 inserts into a hydrophobic pocket formed by Gi residues L194S3.01, F336H5.08, I343H5.15, and I344H5.16, or mGαi/s/q residues F228H5.08, I235H5.15, and L236H5.16, stabilizing a significant hydrophobic environment. c, Structural comparison of the α5 and αN helices in the TUG-1375-AZ-1729-FFA2-Gi (yellow), TUG-1375-compound187-FFA2-miniGq (yellow green), and TUG-1375-compound-FFA2-Gi (pink) complexes. The α5 helix adopts a highly similar conformation across all three structures, whereas the αN helix exhibits greater variability. d, Polar interactions between FFA2 and the α5 helix of miniGq (right panel), which are missing in the structures with Gi (left panel). Polar interactions are shown as black dashed lines.
Extended Data Fig. 7 Compound3 and ligand selectivity at Site 2.
a, Effect of various concentrations of compound3 on the concentration-response profiles of 4-CMTB and C3. Assays were performed as described in Methods, with the following adjustments: (1) 2,000 cells were seeded per well; (2) cAMP accumulation was induced by 0.1 µM forskolin; (3) cells were incubated for 45 min with indicated agonists. Error bars represent mean ± s.e.m. from 3 biologically independent experiments, each performed in triplicate. p-values refer to changes in potency and are reported according to one-way ANOVA test with Dunnet’s post hoc analysis. b, Comparison of Site 2 in FFA2 and similar pockets in FFA1, FFA3, and FFA4. AZ-1729 and compound187 were docked to the other FFAs by aligning their structures to FFA2 structures with these two PAMs. Severe steric clashes as indicated by red circles are observed between AZ-1729 or cmpd187 and other FFAs, indicating a high selectivity of these two PAMs for FFA2. c, Comparison of Site 2 in FFA2 and the similar allosteric site in C5aR. Left: FFA2 (blue) in complex with the PAM AZ-1729 (orange); Middle: FFA2 (green) in complex with the PAM compound187 (pink); Right: C5a receptor (C5aR, brown) in complex with the NAM avacopan (light blue). The crystal structure of the C5aR-avacopan complex (PDB ID: 6C1R) illustrates the binding mode of this allosteric antagonist.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figs. 1–3, Supplementary Tables 1–3 and Supplementary References.
Supplementary Data
The zip folder contains source data for the main figures and the extended data figures.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Guseinov, AA., Jenkins, L. et al. Allosteric modulation and biased signalling at free fatty acid receptor 2. Nature 643, 1428–1438 (2025). https://doi.org/10.1038/s41586-025-09186-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09186-6
This article is cited by
-
Molecular insights into ago-allosteric modulation at cysteinyl leukotriene receptor 2
Nature Communications (2025)