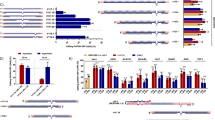
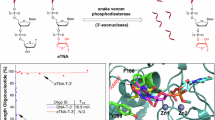
Abstract
Therapeutic small interfering RNA (siRNA) requires sugar and backbone modifications to inhibit nuclease degradation. However, metabolic stabilization by phosphorothioate (PS), the only backbone chemistry used clinically, may be insufficient for targeting extrahepatic tissues. To improve oligonucleotide stabilization, we report the discovery, synthesis and characterization of extended nucleic acid (exNA) consisting of a methylene insertion between the 5′-C and 5′-OH of a nucleoside. exNA incorporation is compatible with common oligonucleotide synthetic protocols and the PS backbone, provides stabilization against 3′ and 5′ exonucleases and is tolerated at multiple oligonucleotide positions. A combined exNA–PS backbone enhances resistance to 3′ exonuclease by ~32-fold over the conventional PS backbone and by >1,000-fold over the natural phosphodiester backbone, improving tissue exposure, tissue accumulation and efficacy in mice, both systemically and in the brain. The improved efficacy and durability imparted by exNA may enable therapeutic interventions in extrahepatic tissues, both with siRNA and with other oligonucleotides such as CRISPR guide RNA, antisense oligonucleotides, mRNA and tRNA.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data supporting the findings of this study are available from the corresponding authors upon reasonable request. The raw NMR data are included in the Supplementary Information.
References
Khvorova, A. & Watts, J. K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 35, 238–248 (2017).
Crooke, S. T. et al. RNA targeted therapeutics. Cell Metab. 27, 714–739 (2018).
Levin, A. A. Treating disease at the RNA level with oligonucleotides. N. Engl. J. Med. 380, 57–70 (2019).
Shen, X. & Corey, D. R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 46, 1584–1600 (2018).
Egli, M. & Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 51, 2529–2573 (2023).
Chen, Y. et al. Delivery of therapeutic small interfering RNA: the current patent-based landscape. Mol. Ther. Nucleic Acids 29, 150–161 (2022).
Egli, M. & Manoharan, M. Re-engineering RNA molecules into therapeutic agents. Acc. Chem. Res. 52, 1036–1047 (2019).
Biscans, A. et al. The chemical structure and phosphorothioate content of hydrophobically modified siRNAs impact extrahepatic distribution and efficacy. Nucleic Acids Res. 48, 7665–7680 (2020).
Gökirmak, T. et al. Overcoming the challenges of tissue delivery for oligonucleotide therapeutics. Trends Pharmacol. Sci. 42, 588–604 (2021).
Schlegel, M. K. et al. From bench to bedside: improving the clinical safety of GalNAc–siRNA conjugates using seed-pairing destabilization. Nucleic Acids Res. 50, 6656–6670 (2022).
Mukherjee, D. et al. Analysis of RNA exonucleolytic activities in cellular extracts. Methods Mol. Biol. 257, 193–212 (2004).
Nair, J. K. et al. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc–siRNA conjugates. Nucleic Acids Res. 45, 10969–10977 (2017).
Foster, D. J. et al. Advanced siRNA designs further improve in vivo performance of GalNAc–siRNA conjugates. Mol. Ther. 26, 708–717 (2018).
Brown, C. R. et al. Investigating the pharmacodynamic durability of GalNAc–siRNA conjugates. Nucleic Acids Res. 48, 11827–11844 (2020).
Allison, H. et al. RNA interference using boranophosphate siRNAs: structure–activity relationship. Nucleic Acids Res. 32, 5991–6000 (2004).
Allison, H. et al. High potency silencing by single-stranded boranophosphate siRNA. Nucleic Acids Res. 34, 2773–2781 (2006).
Maede, B. et al. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat. Biotechnol. 32, 1256–1261 (2014).
Hardcastle, T. et al. A single amide linkage in the passenger strand suppresses its activity and enhances guide strand targeting of siRNAs. ACS Chem. Biol. 13, 533–536 (2018).
Richter, M. et al. Amide modifications in the seed region of the guide strand improve the on-target specificity of short interfering RNA. ACS Chem. Biol. 18, 7–11 (2023).
Janas, M. M. et al. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 9, 723 (2018).
Matsuda, S. et al. The α-(l)-threofuranosyl nucleic acid modification improves stability, potency, safety, and Ago2 binding and mitigates off-target effects of small interfering RNAs. J. Am. Chem. Soc. 145, 19691–19706 (2023).
Lima, W. F. et al. Single-stranded siRNAs activate RNAi in animals. Cell 150, 883–894 (2012).
Parmar, R. et al. 5′-(E)-Vinylphosphonate: a stable phosphate mimic can improve the RNAi activity of siRNA–GalNAc conjugates. ChemBioChem 17, 985–989 (2015).
Elkayam, E. et al. siRNA carrying an (E)-vinylphosphonate moiety at the 5′-end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 45, 3528–3536 (2017).
Haraszti, R. A. et al. 5′-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 43, 2993–3011 (2015).
Yamada, K. et al. Structurally constrained phosphonate internucleotide linkage impacts oligonucleotide–enzyme interaction, and modulates siRNA activity and allele specificity. Nucleic Acids Res. 49, 12069–12088 (2021).
Kupryushkin, M. S. et al. Phosphoryl guanidines: a new type of nucleic acid analogues. Acta Naturae 6, 116–118 (2014).
Liu, W. et al. Impact of stereopure chimeric backbone chemistries on the potency and durability of gene silencing by RNA interference. Nucleic Acids Res. 51, 4126–4147 (2023).
Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 24, 374–387 (2014).
Schirle, N. T. et al. Structural analysis of human Argonaute-2 bound to a modified siRNA guide. J. Am. Chem. Soc. 138, 8694–8697 (2016).
Fitzgerald, K. et al. A highly durable RNAi therapeutic inhibitor of PCSK9. N. Engl. J. Med. 376, 41–51 (2017).
Kim, J. et al. Metabolite profiling of the antisense oligonucleotide eluforsen using liquid chromatography–mass spectrometry. Mol. Ther. Nucleic Acids. 17, 714–725 (2019).
Li, J. et al. Nonclinical pharmacokinetics and absorption, distribution, metabolism, and excretion of Givosiran, the first approved N-acetylgalactosamine-conjugated RNA interference therapeutic. Drug Metab. Dispos. 49, 572–580 (2021).
Moore, L. D. et al. DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38 (2013).
Mikhailov, S. N. et al. Use of 5-deoxy-ribo-hexofuranose derivatives for the preparation of 5′-nucleotide phosphonates and homoribonucleosides. Collect. Czech. Chem. Commun. 54, 1055–1066 (1989).
Robins, M. J. et al. Biomimetic modeling of the decomposition of 2′-chloro-2′-deoxynucleotides by ribonucleotide reductases to give 3(2H)-furanones which can effect mechanism-based inactivation by Michael-type alkylation. J. Am. Chem. Soc. 118, 11317–11318 (1996).
Haly, B. et al. An extended phosphate linkage: synthesis, hybridization and modeling studies of modified oligonucleotides. Nucleosides Nucleotides 15, 1383–1395 (1996).
Khvorova, A., Roux, L. M. & Yamada, K. Modified oligonucleotides with increased stability. US Patent 2020025017-W (2020); https://portal.unifiedpatents.com/patents/patent/WO-2020198509-A3
Khvorova, A., Roux, L. M. & Yamada, K. Synthetic oligonucleotides having regions of block and cluster modifications. US Patent 20210395739A1 (2021); https://patents.google.com/patent/US20210395739A1/en
Khvorova, A., Roux, L. M. & Yamada, K. Synthesis of modified oligonucleotides with increased stability. US Patent 20220010309A1 (2022); https://patents.google.com/patent/US20220010309A1/en
Traube, F. R. et al. The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol. 14, 1099–1107 (2017).
Liu, R. et al. Methylation across the central dogma in health and diseases: new therapeutic strategies. Signal Transduct. Target. Ther. 8, 310 (2023).
Kachare, D. et al. Phospho-carboxylic anhydride of a homologated nucleoside leads to primer degradation in the presence of a polymerase. Bioorg. Med. Chem. 24, 2720–2723 (2014).
Kel’in, A. V. et al. Structural basis of duplex thermodynamic stability and enhanced nuclease resistance of 5′-C-methyl pyrimidine-modified oligonucleotides. J. Org. Chem. 81, 2261–2279 (2016).
Sinha, N. D. et al. β-Cyanoethyl N,N-dialkylamino/N-morpholinomonochloro phosphoramidites, new phosphitylating agents facilitating ease of deprotection and work-up of synthesized oligonucleotides. Tetrahedron Lett. 24, 5843–5846 (1983).
Clavé, G. et al. Modified internucleoside linkages for nuclease-resistant oligonucleotides. RSC Chem. Biol. 2, 94–150 (2021).
Schirle, N. T. & MacRae, I. J. The crystal structure of human Argonaute2. Science 336, 1037–1040 (2012).
Song, J. J. et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10, 1026–1032 (2003).
Elkayam, E. et al. siRNA carrying an (E)-vinylphosphonate moiety at the 5′ end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 45, 3528–3536 (2017).
Ma, J. B. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429, 318–322 (2004).
Alterman, J. F. et al. Hydrophobically modified siRNAs silence Huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids 4, e266 (2015).
Hassler, M. R. et al. Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res. 46, 2185–2196 (2018).
Salomon, W. E. et al. Single-molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell 162, 84–95 (2015).
Nakanishi, K. et al. Structure of yeast Argonaute with guide RNA. Nature 486, 368–374 (2012).
Klum, S. M. et al. Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J. 37, 75–88 (2018).
Kobayashi, Y. siRNA seed region is divided into two functionally different domains in RNA interference in response to 2′-OMe modifications. ACS Omega 7, 2398–2410 (2022).
Shmushkovich, T. et al. Functional features defining the efficacy of cholesterol-conjugated, self-deliverable, chemically modified siRNAs. Nucleic Acids Res. 46, 10905–10916 (2018).
Chu, Y. L. et al. siRNA function in RNAi: a chemical modification analysis. RNA 9, 1034–1048 (2003).
Zheng, J. Single modification at position 14 of siRNA strand abolishes its gene-silencing activity by decreasing both RISC loading and target degradation. FASEB J. 27, 4017–4026 (2013).
Haley, B. & Zamore, P. D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 11, 599–606 (2004).
Ameres, S. L. et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–1539 (2020).
De, N. et al. Highly complementary target RNAs promote release of guide RNAs from human Argonaute2. Mol. Cell. 50, 344–355 (2013).
Kumar, P. et al. Chimeric siRNAs with chemically modified pentofuranose and hexopyranose nucleotides: altritol-nucleotide (ANA) containing GalNAc-siRNA conjugates: in vitro and in vivo RNAi activity and resistance to 5′-exonuclease. Nucleic Acids Res. 48, 4028–4040 (2020).
Shaw, J.-P. et al. Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res. 19, 747–750 (1991).
Davis, S. M. et al. 2′-O-Methyl at 20-mer guide strand 3′ termini may negatively affect target silencing activity of fully chemically modified siRNA. Mol. Ther. Nucleic Acids. 21, 266–277 (2020).
Choung, S. et al. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 342, 919–927 (2006).
Biscans, A. et al. Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Res. 47, 1082–1096 (2019).
Roehl, I., Schuster, M. & Seiffert, S. Oligonucleotide detection method. US Patent 20110201006-A1 (2011); https://portal.unifiedpatents.com/patents/patent/US-20110201006-A1
Alterman, J. F. et al. A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat. Biotechnol. 37, 884–894 (2019).
Slow, E. J. et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 12, 1555–1567 (2003).
Orans, J. et al. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 145, 212–223 (2011).
Jinek, M. et al. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell 41, 600–608 (2011).
Brautigam, C. A. et al. Structural principles for the inhibition of the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol. 277, 363–377 (1998).
Dowdy, S. F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 35, 222–229 (2017).
Springer, A. D. & Dowdy, S. F. GalNAc-siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 28, 109–118 (2018).
Laursen, M. B. et al. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol. Biosyst. 6, 862–870 (2010).
Aiba, Y. et al. Allele-selective inhibition of expression of huntingtin and ataxin-3 by RNA duplexes containing unlocked nucleic acid substitutions. Biochemistry 52, 9329–9338 (2013).
Bartel, D. P. Metazoan microRNAs. Cell 173, 20–51 (2018).
Zhu, Y. et al. RNA-based therapeutics: an overview and prospectus. Cell Death Dis. 13, 644 (2022).
Dolgin, E. tRNA therapeutics burst onto startup scene. Nat. Biotechnol. 40, 283–286 (2022).
Barrangou, R. & Doudna, J. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941 (2016).
Patutina, O. A. et al. Mesyl phosphoramidate backbone modified antisense oligonucleotides targeting miR-21 with enhanced in vivo therapeutic potency. Proc. Natl Acad. Sci. USA 117, 32370–32379 (2020).
Nikan, M. et al. Synthesis and evaluation of parenchymal retention and efficacy of a metabolically stable O-phosphocholine-N-docosahexaenoyl-l-serine siRNA conjugate in mouse brain. Bioconjug. Chem. 28, 1758–1756 (2017).
Westmanu, E. et al. Removal of t-butyldimethylsilyl protection in RNA-synthesis. Triethylamine trihydrofluoride (TEA, 3HF) is a more reliable alternative to tetrabutylammonium fluoride (TBAF). Nucleic Acids Res. 22, 2430–2431 (1994).
Godinho, B. M. D. C. et al. Pharmacokinetic profiling of conjugated therapeutic oligonucleotides: a high-throughput method based upon serial blood microsampling coupled to peptide nucleic acid hybridization assay. Nucleic Acid Ther. 6, 323–334 (2017).
Coles, A. H. et al. A high-throughput method for direct detection of therapeutic oligonucleotide-induced gene silencing in vivo. Nucleic Acid Ther. 26, 86–92 (2016).
Peng, S. X. et al. Improved pharmacokinetic and bioavailability support of drug discovery using serial blood sampling in mice. J. Pharm. Sci. 98, 1877–1884 (2009).
Conroy, F. et al. Chemical engineering of therapeutic siRNAs for allele-specific gene silencing in Huntington’s disease models. Nat. Commun. 13, 5802 (2022).
DiFiglia, M. et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron 14, 1075–1081 (1995).
Brooks, M. E. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 9, 378–400 (2017).
Acknowledgements
This project was funded by the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (R01 NS104022 to A.K.), S10 (OD020012 to A.K.), Maximizing Investigators’ Research Award (R35 GM131839 to A.K.), NIH CREATE (U01 NS114098 to N.A.), Cure Huntington’s Disease Initiative Foundation (RecID A-5038 to N.A.) and the Berman–Topper Fund (to A.K. and N.A.). The authors would like to thank the University of Massachusetts Chan Medical School Animal Medicine Department and veterinary technicians for contributing to the large-animal studies. Some icons in Figs. 5b and 6b were adapted from BioRender.com. The authors would also like to thank E. Haberlin for proofreading the manuscript, A. Ali for maintaining the infrastructure for nuclear magnetic resonance (NMR) and Y. Tan and S. Nguyen for measurement of high-resolution mass analysis.
Author information
Authors and Affiliations
Contributions
K.Y. and A.K. conceived the project. K.Y., V.N.H. and A.K. contributed to the experimental design for exNA studies in hepatic and extrahepatic tissues. K.Y., V.N.H., J.C., H.H.F., Q.T., A.B., R.C.F, A.S., C.L. and B.M.D.C.G. conducted the experiments. C.M.F., K.Y., R.M., E.S., J.F.A., M.D., N.A. and A.K. contributed to the experimental design for exNA studies in CNS. C.M.F., R.M., E.S. and J.D.P conducted experiments. K.Y. and N.Y. contributed to the exNA thermal stability and nuclease resistance study. K.Y., N.Y., B.M.B., N.M., S.O.J. and D.E. contributed to the oligonucleotide synthesis. S.H. contributed to the statistical analysis of exNA CNS studies. V.N.H. and M.R.H. gave inspirational intellectual contributions. K.Y. and A.K. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
K.Y., V.N.H. and A.K. have filed patent applications for exNA platforms. A.K. discloses ownership of stocks in RXi Pharmaceuticals and Advirna, and is a founder of Atalanta Therapeutics and Comanche Biopharma. V.N.H. is an employee of Comanche Biopharma and owns stock options. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Eriks Rozners and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Synthesis of exNA phosphoramidites (6a and 6b) and exNA-modified oligonucleotide.
Reagent and conditions are as follows: (i) IBX/CH3CN, 85 °C, 1.5–2 h. (ii) CH3PPh3Br, tBuOK, THF, 0 °C then rt, overnight, 2a: 75% (2 steps), 2b: 67% (2 steps). (iii) 9-BBN, THF, 0 °C, overnight. (iv) NaBO3·4H2O, MeOH, THF, H2O, 0 °C then rt, overnight, 3a: 62% (2 steps), 3b: N.D. (v) DMTr-Cl, pyridine, rt, 2 h. (vi) 0.1 M TBAF, THF, rt, 1 h, 5a: 93% (2 steps), 5b: 12% (3 steps). (vii) 2-Cyanoethyl N,N-diisopropylchlorophosphoramidite, DIPEA, CH2Cl2, 0 °C then rt, 0.5 h, 6a: 86%, 6b: 81%. (viii) RNA synthesizer (Supplementary Information).
Supplementary information
Supplementary information
Supplementary Note (general remarks), Figs. 1–9, Tables 1–11, Scheme 1 and Data (synthetic oligonucleotide sequence information, synthesis procedures of compounds, LC–MS data and all raw NMR material data).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yamada, K., Hariharan, V.N., Caiazzi, J. et al. Enhancing siRNA efficacy in vivo with extended nucleic acid backbones. Nat Biotechnol 43, 904–913 (2025). https://doi.org/10.1038/s41587-024-02336-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41587-024-02336-7
This article is cited by
-
Development and evaluation of siRNA-mediated gene silencing strategies for ADO2 therapy utilizing iPSCs model and DMPC-SPIONs delivery system
Stem Cell Research & Therapy (2025)
-
Stereo-random oligonucleotides enable efficient recruitment of ADAR in vitro and in vivo
Nature Communications (2025)
-
Chemical strategies for brain delivery of genomic therapy
Nature Reviews Chemistry (2025)
-
An epimer of threose nucleic acid enhances oligonucleotide exonuclease resistance through end capping
Communications Chemistry (2025)
-
RNA chemistry and therapeutics
Nature Reviews Drug Discovery (2025)