Abstract
Unlike sequencing-based methods, which require cell lysis, optical pooled genetic screens enable investigation of spatial phenotypes, including cell morphology, protein subcellular localization, cell–cell interactions and tissue organization, in response to targeted CRISPR perturbations. Here we report a multimodal optical pooled CRISPR screening method, which we call CRISPRmap. CRISPRmap combines in situ CRISPR guide-identifying barcode readout with multiplexed immunofluorescence and RNA detection. Barcodes are detected and read out through combinatorial hybridization of DNA oligos, enhancing barcode detection efficiency. CRISPRmap enables in situ barcode readout in cell types and contexts that were elusive to conventional optical pooled screening, including cultured primary cells, embryonic stem cells, induced pluripotent stem cells, derived neurons and in vivo cells in a tissue context. We conducted a screen in a breast cancer cell line of the effects of DNA damage repair gene variants on cellular responses to commonly used cancer therapies, and we show that optical phenotyping pinpoints likely pathogenic patient-derived mutations that were previously classified as variants of unknown clinical significance.
Similar content being viewed by others
Main
Pooled CRISPR screens, where the responses of many individual cells to different genetic perturbations can be measured in parallel, are enabling an increasing variety of high-throughput genetic analyses. All such studies must correlate phenotype with the specific genetic modification, which is generally identified by the readout of a DNA-encoded barcode. This was originally achieved for assays of cell viability or expression of a marker used for cell sorting by measuring enrichment or depletion of specific barcodes from the bulk or sorted cell population. To allow screening for molecular phenotypes, it is necessary to measure these parameters and associated barcodes in single cells. Coupling single-cell RNA sequencing (scRNA-seq) to pooled CRISPR screens has vastly expanded our ability to study the transcriptomic response to perturbations1,2, but it suffers from the necessity to isolate and lyse cells. As such, scRNA-seq approaches are agnostic to spatial organization of intercellular and intracellular phenotypes. Imaging techniques have emerged to enhance these screens3,4,5,6, capturing complex cellular behaviors and dynamic phenotypic changes, including intricate cellular morphology and molecular distribution, without destroying the cells. These advancements make it possible to observe a wide array of spatially resolved cellular phenotypes in genetic screens.
The integration of single-cell multimodal profiling, which concurrently analyzes proteins and RNA, is crucial for a nuanced understanding of cellular function. Although RNA profiling provides data on gene expression, sole reliance upon RNA profiling can be fraught with incomplete or erroneous conclusions due to post-transcriptional and post-translational processes that RNA sequencing is blind to. Combining multimodal profiling with the ability of optical pooled CRISPR screens to perturb at a pathway-wide or even a genome-wide scale has not yet been widely adopted, despite its potential to expand understanding of how cellular pathways are regulated in health and disease states, such as cancer.
Pooled base editor screens have recently emerged as a powerful method for mutational scanning, enabling researchers to directly alter endogenous proteins within live cells and, thereby, revolutionize the study of proteins in their natural environments7,8. These screens can use nuclease hybrids of deficient Cas9 with APOBEC1 (BE3) guided by single guide RNAs (sgRNAs) to induce specific point mutations through direct chemical modification, offering a precise means of editing9. These precise base changes largely occur within a defined genomic nucleotide window10. The precision of editing is advantageous for high-resolution analysis of protein function and the mapping of sequence–activity relationships.
Leveraging multimodal optical pooled screens to interrogate key cellular and clinically relevant pathways holds immense potential. Indeed, as next-generation sequencing (NGS) has become more common in clinical oncology, there has been an increasing number of variants of unknown significance (VUSs) in genes linked to cancer predisposition and aggressiveness11. In particular, VUSs are commonly identified in DNA damage response (DDR) genes, which are critical for genomic stability, DNA damage signaling, DNA damage-related checkpoints and DNA repair genes12. The importance of understanding whether a VUS is functional or a passenger event is underscored by real-world clinical consequences. For example, patients with particular VUSs may be candidates for therapy leveraging impairment of a DDR pathway, or relatives may be at elevated cancer risk if such a mutation arises in the germline. As such, understanding the normal function of each gene and how key mutations alter homeostasis is critical. To disentangle the phenotypic difference among these variants during DNA damage repair, we applied five different treatments to MCF7 breast cancer cells to introduce DNA damage through different mechanisms of action. Ionizing irradiation directly introduces DNA double-strand breaks to the target cells. Camptothecin specifically inhibits the DNA topoisomerase I, causing replication fork collisions13. Olaparib targets the poly(ADP‐ribose) polymerase to blockade the repair of single‐strand DNA breaks, which results in DNA double-strand breaks during replication14. Cisplatin causes inter-strand crosslinks by crosslinking the purine bases on the DNA15. Etoposide introduces DNA double-strand breaks by targeting topoisomerase II (ref. 16). These DNA-damaging agents are used clinically, and the variant-specific responses to drugs thus holds potential to help prioritize therapeutic strategies. Furthermore, due to the essential nature of many DDR genes for cell viability, efforts to genetically deplete DDR genes may not recapitulate clinically observed variants and function. Hence, efforts to interrogate the function of these proteins with point mutations are necessary to understand their function.
Ideally, the effects of variants, or any genomic perturbation, would be studied in the native context that the cell encounters in vivo. Recent technological advancements have allowed for protein epitope-based identification of CRISPR guide expressions within tumor tissues at a single-cell resolution17. RNA-based barcoding holds the promise to increase the complexity of these libraries, but its application in tissue contexts has not yet been reported.
Building on these innovations, we developed a sequencing-free barcode readout approach for optical pooled CRISPR screens that is compatible with highly multiplexed antibody and RNA transcript profiling. We applied this method to a breast cancer cell line to evaluate how 292 nucleotide variants across 27 key DDR genes affect the DDR by visualizing the recruitment of DDR proteins to sites of DNA damage during different cell cycle phases after ionizing radiation exposure. Our work also demonstrates the capability to optically read RNA-encoded barcodes in tissue sections, linked with multiplexed antibody detection. This serves as a stepping stone toward in vivo CRISPR screens that can map the cellular landscape and pathway behaviors at a subcellular level.
Results
CRISPRmap enables optical readout of cellular barcodes
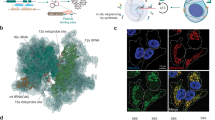
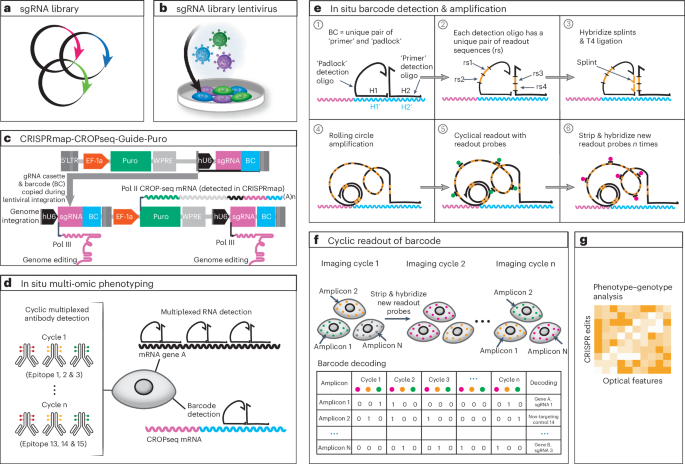
Pooled CRISPR screens typically introduce a single perturbation and its corresponding barcode in a cell through lentiviral infection at a low multiplicity of infection (MOI) (Fig. 1a,b). Our barcode is expressed as part of an abundant mRNA encoding for a selection marker4 (Fig. 1c). In CRISPRmap, the cellular barcode consists of a unique combination of two adjacent 30-bp hybridization sequences. The first step of barcode detection occurs through hybridization of a pair of single-stranded DNA oligos that are complementary to the adjacent hybridization sequences on the transcript18 (Fig. 1e). In our approach, the primer and padlock oligos each contain a unique pair of 20mer readout sequences. Collectively, the four 20mer sequences form a unique combinatorial readout set. Padlock probe circularization by T4 DNA ligase is dependent on hybridization of splint oligos, which bind to the 20mers on the primer oligo (Fig. 1e). Subsequently, rolling circle amplification (RCA) is initiated through the primer oligo. Crucially, valid amplicons rely on AND logic for the primer, padlock and both splint oligos. The readout set, and, thus, by extension, the cellular barcode, is identified by cyclical hybridization rounds with dye-conjugated oligos (readout probes)19,20. Distributing the readout set over primer and padlock probes enables us to identify improperly self-ligated padlock oligos (readout set lacks primer readouts), or unallowed primer–padlock pairing (invalid readout set), and exclude them from analysis. Our cyclical hybridization readout approach was designed to minimize dependence on third-party sequencing reagents, tissue degradation during cyclic enzymatic steps and reagent cost of the assay (Supplementary Table 1).
a, Synthesized sgRNA and barcode library are cloned onto the modified CROPseq vector for sgRNA and barcode expression. b, Plasmids are lentivirally transduced into target cells. c, Design of the CRISPmap-CROPseq-Guide-Puro vector. Human U6 (hU6) promoter (black) drives the sgRNA expression by RNA Pol III, and a Pol III stop signal is inserted between the sgRNA and the barcode. The hU6-sgRNA-stop cassette and the barcode are inserted in the 3′ LTR sequence and will, thus, be copied during genome integration to the upstream of the EF-1a promoter. The EF-1a promoter drives the expression of the CROPseq mRNA by RNA Pol II, which expresses the puromycin resistance gene (green), hU6 (black), sgRNA (magenta) and barcode (cyan). This figure was adapted from Datlinger et al.1. d, In situ multimodal phenotyping and CRISPRmap barcode detection. Multimodal phenotyping interrogates proteomic and transcriptomic states, and CRISPRmap barcode readout identifies the sgRNA identity. Cyclic antibody staining (IBEX) is used to detect dozens of epitopes. Pairs of padlock and primer oligos are hybridized to the CROPseq mRNA or endogenous RNAs to detect CRISPRmap barcodes or target RNA transcripts. e, In situ barcode detection and amplification. Padlock and primer oligos hybridize to the barcode sequence on the CROPseq mRNA. Padlock and primer each encode a unique pair of readout sequences. Splints hybridize to the corresponding readout sequences on primer oligos. Padlock oligos and splints are joined by T4 ligation to enable RCA. Fluorophore-conjugated readout probes hybridize to readout sequences on the amplicons in a cyclic manner for barcode identification. f, Barcode readout and decoding. Images across fluorescence channels and imaging cycles are co-registered into a unified readout stack. Barcode decoding at the amplicon level is achieved through spot detection, assigning a bit code (0 for absence, 1 for presence) in each image to generate a barcode across images. If the barcode aligns with a guide-identifying barcode in the codebook, a guide identity is assigned to the corresponding amplicon. g, Phenotype–genotype analysis. Multimodal and multiplexed phenotyping provides high-dimensional optical features for systematic analysis. BC, barcode; LTR, long terminal repeat; Pol, polymerase.
To develop and optimize our approach, we transduced a small pilot lentiviral library containing five green fluorescent protein (GFP)-targeting CRISPR guides and five non-targeting CRISPR guides in a HT1080–Cas9 cell line expressing copGFP. We performed lentiviral library preparation (Methods) and infected cells at MOI < 0.1 to ensure that most infected cells will be edited by a single guide and express a single barcode after puromycin selection. To couple our optical phenotype (GFP expression) to the CRISPR edit, we performed CRISPRmap (Methods and Fig. 2a). For this small pilot experiment, the readout set was imaged in two channels over four imaging cycles; larger libraries discussed later were imaged in three channels over eight imaging cycles. Images across all barcode readout cycles and channels were co-registered into an image stack and corrected for global translational shifts (that is, misaligned glass-bottom well plate placement) as well as local translational shifts (that is, cells slightly shifting between imaging rounds). To align the images across all imaging rounds, we calculated the transformation matrices for each round using the TV-L1 implementation of optical flow21 on binary nuclei masks derived from DAPI stains (Methods). In our GFP-targeting CRISPRmap screen, barcode decoding is performed at amplicon level (Methods) by assigning an 8-bit code for each amplicon across the readout cycles and channels, where signal from each readout sequence yields a positive entry (1) and lack of signal yields a negative entry (0) (Fig. 2b). A guide identity (Guide ID) is assigned to an amplicon if the 8-bit code of the amplicon position matches a guide-identifying barcode in the pre-designed library codebook. We found that, of all the amplicons that were positive for four readout probes, 98% coded for an allowed barcode included the library design, whereas 2% of amplicons reported an unallowed barcode (Fig. 2c), despite their relative ratios of 10 of 25 (40%) versus 15 of 25 (60%) possible primer–padlock pairs. In contrast, when we performed CRISPRmap readout on non-barcoded cells, we recovered, on average, 0.2 non-specific barcodes per cell, of which approximately 62% stem from unallowed primer–padlock pairs, in agreement with their relative frequency (Supplementary Table 2). Because CRISPRmap quality control (QC) criteria require at least three barcode spots per cell with two out of three having the same barcode, unspecific binding is unlikely to affect precision of barcode assignment. From a per-cell analysis, we found that, when imaging with a ×20 objective, the median number of guide-assigned amplicons per cell was 11 (Fig. 2d). We restricted further analysis to cells with three or more amplicons and for which the most abundant barcode made up more than two-thirds of the amplicons of the sum of the two most abundant barcodes under a cell segmentation mask. The latter criterion was put in place to retain cells for which imperfect cell segmentation could cover a few amplicons from neighboring cells, causing false association of guide-assigned amplicons to cell masks. With these QC metrics in place, we retained 76% of the cells for further analysis (Fig. 2d and Supplementary Table 2). Finally, we evaluated if we observed the expected optical phenotype for each of the guides in our pilot library and found, indeed, that cells with GFP-targeting guides have significantly lower GFP fluorescence levels than cells with non-targeting control (NTC) guides (Fig. 2c), which, in turn, have similar GFP fluorescence levels as unperturbed cells. Cells with each of the five GFP-targeting guides showed significantly lower GFP levels than cells with any of the NTC guides under pairwise comparison (Supplementary Fig. 1a), thus recapitulating the expected genotype–phenotype relationship. In addition to the ratio of cells that passed QC, we evaluated sensitivity, specificity and precision (Methods) of CRISPRmap in this GFP-pilot dataset, and we found that they compare similarly or more favorably to conventional optical pooled screening (OPS)4 (Supplementary Fig. 1b). These metrics were relatively insensitive to our QC metrics for calling a barcoded cell (Supplementary Fig. 1c–e). Most notable is the decrease in proportion of cells passing QC with increasing minimum number of barcoded amplicons and increasing purity requirements. This effect was less pronounced when plating cells more sparsely (Supplementary Fig. 1d,e), which we attribute to improved cell segmentation. To evaluate the effect of cell segmentation, we performed a ‘barnyard’ experiment where we mixed mTurquoise2+ cells with GFP+ cells, each carrying a unique barcode. We observed excellent separation of the two populations and recovered the expected barcodes (Supplementary Fig. 1f,g), and we obtained high sensitivity, specificity, precision and proportion of cells assigned a barcode (Supplementary Fig. 1b,e).
a, Visualization of genotype–phenotype mapping in cells without (left) or with (middle and right) GFP-pilot library. WGA (magenta) and DAPI (blue) signals are shown (left and middle). Cell boundaries are outlined in blue, whereas decoded barcodes are shown as false-colored spots (magenta for GFP-targeting guides, green for non-targeting guides) (right). Raw GFP signal is displayed by grayscale in all panels. Scale bar, 50 μm. b, Visualization of the barcode readout and phenotyping, showing a cell with a GFP-targeting guide (top row) and a cell with a non-targeting guide (bottom row). Decoded barcodes are displayed as white spots (second-most right column) and projected onto the eight readout images as white circles with raw readout signals displayed in magenta for channel 1 and in green for channel 2 (columns 1–8). Raw GFP signal is shown in grayscale (right-most column). Cell and nuclear boundaries are outlined in blue in all panels. Scale bar, 10 μm (top) and 15 μm (bottom). c, Quantification of all possible primer–padlock combinations, showing robust detection of the 10 allowed combinations and minimal detection of the 15 unallowed combinations. d, Distribution of the number of assigned amplicons per cell under the standard QC (Methods). e, Quantification of genotype–phenotype mapping showing that cells with GFP-targeting guides have significantly reduced GFP fluorescence (P = 1.53 × 10−209). Two-sided Mann–Whitney test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. n = 1 with 4,620 transduced cells, n = 1 with 4,810 non-transduced cells. Boxes indicate the median and interquartile range (IQR) with whiskers extending 1.5× IQR past the upper and lower quartiles. f, Barcode detection between conventional OPS and CRISPRmap for HT1080 cells, fibroblasts, iPSCs, iMNs and hESCs. Scale bar, 10 μm. g, Fraction of cells with barcode detection in CRISPRmap on fibroblasts (n = 2 biological replicates), HT1080 cells (n = 5 from four biological replicates), iPSCs (n = 5 from two biological replicates), iMNs and hESCs and conventional OPS on fibroblasts (n = 3 from two biological replicates), HT1080 cells, iPSCs, iMNs and hESCs. n = 2 technical replicates unless otherwise specified. Data are presented as mean values ± 95% confidence interval (CI). BC, barcode.
To assess guide representation throughout library preparation, infection and optical readout of the barcodes, we performed NGS on polymerase chain reaction (PCR) product of the amplified synthesized DNA oligonucleotide pool, the CRISPRmap-CROPseq plasmid pool and the genomic DNA from the cells transduced with the library. We further compared the sequencing result to the relative guide abundance of cells with optically identified guide identities, and we observed highly correlated guide frequencies among all of these stages (Supplementary Fig. 1h,i). Moreover, analysis showed that there was minimal recombination that decoupled the sgRNA from its intended barcode during library preparation. Sequencing of genomic DNA from infected cells revealed that 93% of the reads had a perfect match between sgRNA and CRISPRmap barcode (Supplementary Fig. 1j), whereas 4% showed recombination such that a guide recombined with a different barcode in the pool, to which our optical readout is agnostic. Another 2% lost the barcode and would, thus, not be detected optically. Very few reads had either no guide or an unallowed barcode.
Interestingly, CRISPRmap barcode decoding at the amplicon level led to the detection of some double-transduced cells. These cells express two barcodes, each with a unique spatial pattern (Supplementary Fig. 1k). Performing a series of experiments with increasing MOI indeed observed an increase of cells with double infections (Supplementary Fig. 1l and Supplementary Table 2), albeit a smaller increase than expected from Poisson statistics due to experimental conditions (Methods). Future studies could leverage this feature to study genetic interactions through combinatorial perturbations by infecting pooled libraries at a higher MOI.
CRISPRmap barcodes primary fibroblasts, human embryonic stem cells, induced pluripotent stem cells and motor neuron cells
To evaluate CRISPRmap outside the context of immortalized or cancer cell lines (Supplementary Fig. 2), we profiled primary fibroblasts, human embryonic stem cells (hESCs), induced pluripotent stem cells (iPSCs) and motor neurons derived from iPSCs (iMNs) (Fig. 2f). We validated cell type marker (SOX2, OCT4 and NeuN) expression for hESCs, iPSCs and iMNs (Supplementary Fig. 3). Although conventional OPS performed well for HT1080 cells and fibroblasts (Fig. 2f,g and Supplementary Fig. 4), CRISPRmap showed improved barcode detection in the more challenging cell types, such as hESCs, iPSCs and iMNs, recovering more barcode-assigned amplicons per cell and enabling a larger proportion of cells being assigned a barcode (Fig. 2f,g).
Multimodal in situ profiling of perturbed cell states
To unlock the potential of base editor scanning, we aimed to move beyond cellular fitness as a readout and enable detailed characterization of more complex biological processes. Therefore, we sought to combine base editing approaches with optical, single-cell, multiplexed, multimodal approaches, measuring functional responses of dozens of proteins and mRNAs at subcellular resolution. We profiled the proteomic and transcriptomic responses of breast cancer cells to ionizing irradiation, a critical treatment modality for breast cancers, as a function of base-edited variants of 27 core DNA damage repair genes involved in the DDR, homologous recombination and Fanconi anemia (FA) pathways. Specifically, a 364-sgRNA library was lentivirally transduced into an MCF7 cell line expressing BE3 (MCF7-BE3 hereafter)7. Transduced cells were selected in antibiotic-containing medium for 2 d and cultured in antibiotic-free medium for another 2 d, before induction of DNA damage by gamma radiation. Six hours after irradiation, cells were chemically fixed and profiled for protein expression, mRNA expression and CRISPR barcode readout (Fig. 3a,b and Supplementary Fig. 5). In this dataset, we profiled 226,369 single cells that met all barcode calling quality metrics, resulting in an average coverage of 310 cells per variant in the library in each experimental condition.
a, Experimental workflow (Methods). Image was made using BioRender. b, Subcellular distribution of six DDR protein stains (top row), five cell cycle regulator stains (middle row), barcode detection (middle row, most right) and transcript detection for six genes (bottom row) for a single cell. Cell and nuclear segmentation are outlined in blue, raw antibody signal and transcript detection in grayscale. Decoded barcodes are shown as false-colored (magenta) spots. Only data under the cell segmentation mask are displayed. Scale bar, 10 μm. c, Quantification of the number of RAD51 foci per cell across cell cycle phases in UNT (n = 1 with 120,253 cells) and IR (n = 1 with 106,116 cells) cells, showing significant foci induction by irradiation and enrichment in the S/G2 phase. Boxes indicate the median and interquartile range (IQR) with whiskers extending 1.5× IQR past the upper and lower quartiles (outliers are omitted). Two-sided Mann–Whitney test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The P values of (G2/S_UNT, G0_IR, G1_IR and M_IR) versus G2/S_IR are 0.00e + 00, 0.00e + 00, 0.00e + 00 and 2.74 × 10−37, respectively. d, As in c for BRCA1 foci. The P values are 0.00e + 00, 0.00e + 00, 0.00e + 00 and 3.78 × 10−15. e, Correlation between RNA-reporting spots per million spots measured by RNAmap and TPM reads from RNA sequencing. Pearson correlation (r) equals 0.84. f, RNA–protein correlation measured by RNAmap and antibody staining for three RNA–protein pairs (Ccna2-cyclin A2, Ccnb1-cyclin B1 and Cdkn1a-p21), showing significantly enriched RNA-reporting spots in cells with high protein expression. Boxes indicate the median and IQR with whiskers extending 1.5× IQR past the upper and lower quartiles. Two-sided Mann–Whitney test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The P values (from left to right) are 0.00e + 00, 0.00e + 00 and 0.00e + 00. g, Visualization of the decoded Ccna2-reporting spots (magenta) and cyclin A2 staining (green). Scale bar, 50 μm. h, As in g for Cdkn1a (magenta) and p21 (green). IR, irradiated; UNT, untreated.
To evaluate how variants alter the cellular response after treatment with ionizing radiation, we applied a recently developed approach, IBEX, which employs a cyclical process of antibody staining and chemical bleaching to facilitate high-resolution imaging of dozens of epitopes within a single sample while preserving its physical integrity22. We visualized a panel of key DDR proteins (RAD51, BRCA1, RPA2, γH2AX, 53BP1 and RAD18), cell cycle phase marker proteins (Ki-67, cyclin A2, cyclin B1 and phospho-histone H3) and apoptosis-related proteins (cleaved PARP1, p21 and p53), and we recovered expected subcellular protein localizations (Fig. 3b) and treatment-specific staining patterns (Fig. 3c,d). We also quantified the micronuclei formation based on DAPI stain (Methods). Accumulation of DDR proteins at damaged genomic loci typically gives rise to punctate immunofluorescence detection patterns (foci) in the nuclei, which we quantify at the single-cell level through automated detection (Methods and Supplementary Fig. 6a,b). Cell cycle–related proteins and transcription factors, on the other hand, are evaluated as average fluorescence across the cellular, nuclear and cytosolic mask (the latter we define as cell mask minus nuclear mask). Comparison of cytosolic to nuclear abundance of a protein allows for quantification of its translocation status. To further assess the specificity of the antibody staining, we compared the expression level and subcellular localization of the measured proteins in gamma-irradiated cells to untreated cells while separating cells into four different cell cycle phases (G0 phase, G1 phase, S/G2 phase and M phase) based on the cell cycle markers that we measured. As expected, we observe significant induction of nuclear foci formation for all the six measured DDR proteins upon gamma irradiation (Fig. 3c,d and Supplementary Fig. 6c–f). In addition, we observed the expected significant enrichment of nuclear foci that function in the homologous recombination pathway (RAD51, BRCA1, RPA2 and RAD18) in the S/G2 phase. Large γH2AX foci are reported to be involved in double-strand break signaling23, and 53BP1 foci thought to promote the non-homologous end-joining pathway showed a slight enrichment in G1 phase over S/G2 phase (Supplementary Fig. 6e,f). Finally, the single-cell and highly multiplexed nature of our data enables us to evaluate the correlation among all optical features that we measured (Supplementary Fig. 6h). As expected, we observed a positive correlation between the nuclear foci involved in homologous recombination (RAD51, BRCA1, RPA2 and RAD18) and the average nuclear intensity of the S/G2-phase markers (cyclin A2 and cyclin B1) and the proliferation marker (Ki-67).
To simultaneously measure the transcriptomic response of cells, we adapted our CRISPRmap barcode detection approach to detect endogenous mRNA transcripts; we call this approach RNAmap (Fig. 1c), which differs from CRISPRmap in three key ways. First, the transcript-hybridizing regions of the primer and padlock detection oligos target adjacent sequences on the endogenous RNA transcripts. The design of gene-specific detection oligos was refined for specificity, a narrow range of melting temperatures (Tm) for primer and padlock oligos, minimal off-target binding and secondary structure (Methods). Second, to promote detection efficiency, we increased the number of primer–padlock oligo pairs to six per RNA transcript. Primer–padlock pairs that share an RNA target also share the same set of readout sequences. Third, to further boost detection efficiency, RNAmap primer detection oligos encode only a single readout sequence, and, as a result, only a single splint oligo needs to undergo ligation to form an RCA template. Consequently, the padlock oligo encodes three readout sequences to enable similar combinatorial readout of transcript identities.
We applied RNAmap to target a panel of 12 genes, selected for their expression in irradiated MCF7-BE3 cells, and to span a range of expression levels. The transcripts profiled include cell cycle–related genes (Ccnb1, Ccna2, Cdkn1a, Cdc20, Kif20a and Cenpe), housekeeping genes (Ppib and Polr2a), DDR-related genes (Ddb2 and Fdxr) and bacterial negative control genes (dapB and fliC) (Fig. 3b). We validated the specificity of detection by comparing optically identified transcript-reporting spots at the population level to bulk RNA sequencing reads, and we observed a Pearson correlation of 0.84 (Fig. 3e). Additional support of specificity is provided by the analysis of the correlation between mRNA and protein expression level for the three cell cycle–related genes. Cells for which we observed high abundance (Supplementary Fig. 6g) of cyclin A2, cyclin B1 and p21 at the protein level have significantly (P < 0.0001, two-sided Mann–Whitney U-test) more corresponding transcripts detected by RNAmap (Fig. 3f–h).
Variant screen recapitulates DDR mechanisms
Building upon our previous work assessing human nucleotide variants across 86 DDR genes and assessing their effect on cell viability7, we selected variants that significantly altered viability in at least one treatment condition in the previous study. We focused the present study on sgRNAs with a single C base in their editing window, to minimize confounding effect of bystander mutations that could obfuscate the phenotypic consequences associated with a guide. Combining the 292 guides targeting DDR genes with 35 guides targeting the AAVS1 safe-harbor site and 37 NTC guides that have minimal targets in the human genome, we applied a 364-guide library (referred as DDR364) to the MCF7-BE3 cells for a multimodal pooled optical base editing screen. Our library includes 162 missense guides, 50 nonsense guides and 80 splice guides (variants that affect splice-donor or splice-acceptor sequences). It is expected that nonsense variants and splice variants are more detrimental to protein function than the missense variants, which are associated with a broader range of effects on protein function. Our library includes 64 variants that the ClinVar database24 annotates as pathogenic/likely pathogenic (P/LP) variants and 75 VUSs. Guide representation of the DDR364 library as sequenced in the plasmid library and recovered by optical barcodes is listed in Supplementary Table 3, and the correlation between guide representation and plasmid library sequencing reads is shown in Supplementary Fig. 7.
We set out to validate if CRISPRmap could recapitulate known phenotypes as a function of specific base edits. First, similar to our previous fitness study7, we found stronger phenotypic changes for guides with higher Rule Set 2 (RS2) on-target efficiency scores, initially created for CRISPR knockout sgRNAs25. Specifically, we first evaluated the abundance in RAD51 foci for variants of genes that are essential for RAD51 foci formation (RAD51D, RAD51C, XRCC3, BRCA1 and BRCA2) relative to negative control guides. Here, we observed a trend of decreasing RAD51 foci with guides with higher RS2 scores, especially for guides that are from the deleterious (nonsense and splice) categories (Fig. 4a). Notably, implementing a minimum RS2 score threshold significantly improved the differentiation between deleterious and negative control guides (two-sided Kolmogorov–Smirnov test, adjusted P (Padj) < 0.0001; Fig. 4c), whereas splice variants below the RS2 threshold showed no statistical significance over control guides (two-sided Kolmogorov–Smirnov test, Padj ≥ 0.05; Fig. 4b). Missense guides targeting these genes generally showed much milder impact on the abundance in RAD51 foci. Similarly, phenotypic changes in BRCA1 foci abundance of BRCA1 variants strongly correlated with RS2 scores (Pearson r = −0.75; Fig. 4d), and cells expressing nonsense or splicing variant guides with RS2 ≥ 0.55 showed significantly fewer BRCA1 foci than cells expressing negative control guides (Fig. 4f). This distinction was not significant for guides with lower RS2 scores (Fig. 4e).
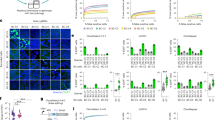
a, Correlation between L2FC in RAD51 foci number and the RS2 on-target score. All guides targeting RAD51 regulators, including RAD51 paralogs (RAD51D, RAD51C, XRCC3), BRCA1 and BRCA2, are shown. Splice and nonsense variants with high RS2 score show more significant L2FC. Pearson correlation (r) = −0.30. b, Quantification of RAD51 foci in irradiated S/G2-phase cells with guides targeting RAD51 regulators that have low RS2 score, grouped by sgRNA category. No or moderate significant separation from cells with control guides was observed. Two-sided Kolmogorov–Smirnov test, *Padj < 0.05, **Padj < 0.01, ***Padj < 0.001, ****Padj < 0.0001. The P values (from top to bottom) are 4.19 × 10−1, 8.03 × 10−3 and 9.79 × 10−1. c, As in b for guides with high RS2 score, showing significant reduction in RAD51 foci in cells with nonsense and splice guides. The P values (from top to bottom) are 9.07 × 10−2, 1.11 × 10−15 and 9.82 × 10−17. d, As in a for L2FC in BRCA1 foci for BRCA1-targeting guides. Pearson correlation (r) = −0.75. e, Same as b for BRCA1 foci and guides targeting BRCA1 that have low RS2 score. The P values (from top to bottom) are 3.52 × 10−1 and 4.99 × 10−1. f, Same as e for guides with high RS2 score, showing significant reduction in BRCA1 foci in all categories. The P values (from top to bottom) are 4.88 × 10−3, 1.30 × 10−10 and 6.86 × 10−6. g, Volcano plot showing no AAVS1-targeting or NTC guides shows statistically significant changes in RAD51 foci. Guides targeting DDR genes with RS2 score ≥ 0.55 and all AAVS1-targeting and NTC guides are shown. Two-sided Kolmogorov–Smirnov test. h, Same as g highlighting guides that result in significant changes in RAD51 foci. i, Gene enrichment analysis of guides causing significant changes in RAD51 foci. One-sided (greater) Fisherʼs exact test. Significance, Padj < 0.05. j, Same as g for BRCA1 foci. k, Same as h for guides that result in significant changes in BRCA1 foci (l) same as i for guides causing significant changes in BRCA1 foci. NS, not significant.
Second, we considered variants to significantly alter nuclear foci formation if there was an absolute log2 fold change (L2FC) > 0.5 in the mean number of foci per cell when compared to the population of negative control guides, and we observed a Benjamini–Hochberg-corrected two-sided Kolmogorov–Smirnov test Padj < 0.05. Based on these criteria, we observed that none of the negative control guides (AAVS1-targeting or NTC guides) significantly altered the number of RAD51 or BRCA1 foci (Fig. 4g,j). Most of the hits significantly reducing the number of RAD51 foci are nonsense or splice variants, annotated in ClinVar as P/LP, but we also identified two missense variants that are annotated as VUSs or not documented (unknown) in the ClinVar database (Fig. 4h). Furthermore, guides that target the BRCA1 and RAD51D genes are significantly enriched in the guides that lead to significant changes in RAD51 foci in the irradiated cells (Fig. 4I), whereas variants that are identified to significantly change the BRCA1 foci (Fig. 4k) are enriched for BRCA1 variants (Fig. 4l), as expected. A BRCA1 missense VUS and an unknown BARD1 missense variant were identified to significantly reduce BRCA1 foci in irradiated cells. The hits identified based on L2FC and Padj thresholds were further confirmed by the measurement of bootstrapped Wasserstein distance (Methods) of each guide from the cells with control guides for RAD51 foci (Supplementary Fig. 8a) and BRCA1 foci (Supplementary Fig. 8b). Analysis of the top four variant hits for RAD51 and BRCA1 (Supplementary Fig. 9a,c, respectively) showed that, on average, those hits would have been distinguished as significant upon screening approximately 60 cells per guide (Supplementary Fig. 9b,d and Methods).
Apart from nuclear foci, we also analyzed protein stains, such as cyclin A2, Ki-67 and p21, for which cells can be categorized into high or low protein expression categories based on the average nuclear fluorescence of the protein stain. Instead of L2FC analysis, for these stains we performed beta-binomial testing (Methods) and observed that three BRCA1 splice variants significantly upregulated the proportion of cells with high p21 expression in the untreated cells (Supplementary Fig. 8c, left), whereas two ATM splice variants reduced the proportion of cells with high p21 expression in irradiated cells (Supplementary Fig. 8c, right).
To further verify perturbation-specific phenotypic changes observed in the pooled screens, we selected two AAVS1-targeting control guides and seven guides targeting BRCA1, BRCA2, BARD1 and PALB2 from different sgRNA and ClinVar categories and with on-target score (RS2) > 0.5 (Supplementary Table 3). BRCA1.234 (splice, P/LP) and BRCA2.207 (Q2580*) (nonsense, P/LP) were identified as hits for RAD51 foci reduction in the pooled screen, whereas BRCA1.234 (splice, P/LP) and BRCA1.416 (H1283Y) (missense, VUS) were identified as hits for BRCA1 foci reduction. We transduced MCF7-BE3 cells with each guide individually and measured the L2FC in RAD51 and BRCA1 foci compared to all control cells in the gamma-irradiated cells. Supplementary Fig. 10 shows representative images for RAD51, BRCA1 and γH2AX foci. We quantified the percentage of cells with more than five RAD51 and BRCA1 foci (Supplementary Fig. 11a,b, respectively) and observed that screen hits transduced as individual guides resulted in a significantly lower percentage compared to the cells transduced with AAVS1-targeting guides, whereas non-screen hits generally showed no or low significance. High correlations of L2FC were observed between guides transduced in the pooled library and those transduced individually (Pearson r = 0.90 for RAD51 foci, Supplementary Fig. 11c; Pearson r = 0.95 for BRCA1 foci, Supplementary Fig. 11d), with hits identified in the pooled screening also showing the most significant changes when transduced individually. In addition, we characterized the base editing efficiency of the three hits that we identified in the pooled screen (BRCA2.207 (Q2580*), BRCA1.234 (splice) and BRCA1.416 (H1283Y)), together with two AAVS1 controls (AAVS1.28 and AAVS1.86). Sanger sequencing of PCR-amplified genomic DNA around the intended editing region for all three hits revealed a high in-window C base editing efficiency (between 50% and 75%), whereas out-of-window C bases typically showed an editing efficiency lower than 25% (Supplementary Fig. 12a).
We also applied the same DDR364 library on MCF7-BE3 cells treated with DNA-damaging agents (camptothecin (CPT); olaparib (OLAP); cisplatin (CISP); etoposide (ETOP)) to study the treatment-specific responses of the gene variants (Supplementary Fig. 8d) with a primary focus on the formation of RAD51 and large γH2AX foci. Similar to the ionizing irradiation screen, we observed that guides targeting the RAD51-relevant genes show more significant loss in RAD51 foci with a higher RS2 score, especially in cells treated with CISP and OLAP (Supplementary Fig. 8e,f). An RS2 score threshold of 0.5 distinguishes deleterious guides that show mild and strong phenotypes. Again, we observed that most of the hits reducing the RAD51 foci in CISP-treated and OLAP-treated cells are splice or nonsense variants, many of which being P/LP (Supplementary Fig. 8g,h), and a significant enrichment of guides targeting RAD51 paralog genes, BRCA1 or BRCA2, can be seen (Supplementary Fig. 8i). A drastic difference in the hits can be seen when comparing the change in large γH2AX foci in ETOP-treated (Supplementary Fig. 8j) and OLAP-treated (Supplementary Fig. 8k) cells. In ETOP-treated cells, we observed that most splice variants of ATM significantly reduced the number of large γH2AX foci, whereas many deleterious variants coming from a mixed background of RAD51-related genes, ATR and FA pathway genes increased large γH2AX foci under other treatments. The enrichment test shows a significant enrichment of FANCA-targeting and FANCI-targeting guides in OLAP-treated cells and ATM-targeting guides in ETOP-treated cells among hits of large γH2AX foci (Supplementary Fig. 8l).
Collectively, these results underscore that CRISPRmap effectively couples barcodes to their corresponding guides and that the expected phenotypic changes are detected by profiling a few hundred cells per variant, despite imperfect efficiencies associated with current base editors. Moreover, CRISPRmap enables the dissection of therapeutic treatment-specific responses of known pathogenic variants and the identification of variants with significant phenotypic effects in the DDR pathways.
Optical screening correlates VUSs with pathogenic variants
Interpreting the functional implications of somatic mutations in cancer, primarily characterized by single-nucleotide changes that often lead to missense VUSs, remains a challenging endeavor. This challenge poses a barrier to effective diagnosis, patient stratification and the management of drug-resistant diseases. Using experimental approaches is crucial in assessing the functional impact of VUSs. This is essential for establishing a link between VUSs and disease-related phenotypes, particularly due to the limited availability of clinical datasets and the infrequent occurrence of certain variants in patient cohorts.
We set out to chart variant effects on the DDR pathways by combining the two CRISPRmap base editing screens that treated cells with ionizing radiation or four DNA-damaging agents commonly used for cancer therapy. In total, we profiled 948,604 cells that passed barcode QC metrics, averaging 372 cells per guide in each treatment condition. We compared benign, VUS, unknown and pathogenic/pathogenic-like variants with our negative control guides, and we found that only benign variants were not significantly different from controls when evaluated on the number of foci features with statistically significant changes from the control cells across all treatment conditions (Supplementary Fig. 8m), whereas P/LP variants led to the highest number of features with significant changes. Moreover, when evaluating the correlation between the L2FC values of optical features between guide pairs, we observed that the distribution of correlations between pairs of sgRNAs that are designed to induce the same variant is higher than the distribution of correlations between all pairwise sgRNAs targeting a particular gene (Supplementary Fig. 8n).
We subsequently clustered variants from functionally related genes based on the optical features measured in the ionizing irradiation–treated and the DNA-damaging agent (CPT, OLAP, CISP and ETOP)–treated cells. When we clustered the variants from the RAD51 paralog genes (RAD51C, RAD51D and XRCC3), we observed a cluster composed of most splice and nonsense variants (Fig. 5b, right cluster). This cluster showed a reduction in RAD51 foci across all treatment conditions and a strong upregulation of γH2AX foci for OLAP and CISP treatments, whereas most missense variants formed a separate cluster with more mild phenotypic changes (Fig. 5b, left cluster).
a, Crucial effectors in DNA damage repair. RAD51 paralogs, including RAD51D, RAD51C and XRCC3, are required for the formation of RAD51 foci at DNA double-strand breaks (DSBs). The BRCA1–BARD1 complex recruits RAD51 to DSB sites. FANCG and FANCI are involved in DNA inter-strand crosslinking (ICL) repair. b, Clustering of guides targeting RAD51 paralog genes (RAD51C, RAD51D and XRCC3), showing a cluster with reduced RAD51 foci in all four DNA-damaging agents–treated and irradiated cells and increased large γH2AX foci in OLAP-treated and CISP-treated cells. The mutations of this cluster are mostly splice and nonsense variants. The left-most column cluster features milder phenotypes mainly associated with missense VUSs. L2FCs in each optical phenotype in corresponding treatment conditions are shown as rows in the heatmap. Cells in all cell cycle phases are included; untreated cells are not included. All guides with RS2 on-target score ≥ 0.5 were included in the clustering and are shown in the heatmap. Columns were cut at a depth of 2, and rows were cut at a depth of 3 based on the dendrogram. Color scale is −1 to 1. c, Same as b for guides targeting BRCA1 and BARD1, showing a cluster with reduced RAD51 and BRCA1 foci in irradiated cells and increased large γH2AX foci and micronuclei in OLAP-treated and CISP-treated cells composed mainly of pathogenic splice and nonsense variants and another cluster with mild phenotypes composed mainly of missense variants. d, Same as b for guides targeting FANCI and FANCG, showing a cluster with mostly splicing variants showing increased large γH2AX foci and micronuclei in OLAP-treated and CISP-treated cells. The left-most column cluster features milder phenotypes mainly associated with missense variants.
Variants of BRCA1 and its heterodimeric binding partner encoding gene BARD1 can also be categorized into two clusters. The right cluster contains most splice and nonsense P/LP variants and shows an expected reduction in RAD51 and BRCA1 foci upon irradiation as well as an increase in large γH2AX foci and micronuclei mostly in OLAP-treated and CISP-treated cells (Fig. 5c). Variants in the left cluster are mostly missense mutations with milder phenotypes. Notably, a missense VUS of BRCA1 (BRCA1.416 (H1283Y)), which renders the H1283Y amino acid change on the BRCA1 protein, clusters with pathogenic variants of BRCA1. Despite being classified as a VUS, a recent study26 classified it to be likely pathogenic based on a BE3 base editing screen with fitness as a readout, and the authors confirmed the loss in cell viability for H1283Y with CRISPR-mediated homology-directed repair. To assess if the missense BRCA1 VUS had a phenotype similar to a nonsense mutation due to a change in protein stability, we performed immunoblotting on cells transduced with individual guides, and we found the missense variant to have full-length protein at similar abundances as the AAVS1 control variant (AAVS1.86), whereas the splice variant (BRCA1.234 (splice)) showed a reduction in full-length protein similar to the small interferring RNA (siRNA)-induced BRCA1 (siBRCA1) knockdown control (Supplementary Fig. 12b). We also performed immunoblotting on two BRCA2 variants to further investigate the protein stability of different types of variants, and we observed a loss of full-length BRCA2 protein in the nonsense variant (BRCA2.207 (Q2580*)) similar to the siRNA-induced BRCA2 (siBRCA2) knockdown control, whereas the missense VUS (BRCA2.438 (A1847T)) had similar full-length protein to the AAVS1 control variant (AAVS1.86) (Supplementary Fig. 12c). Immunoblotting on phospho-KAP1 (pKAP1) indicates the induction of DNA damage upon gamma irradiation, and we observed no correlation between the changes in protein stability and the DNA damage (Supplementary Fig. 12c).
Evaluating variants of Fanconi anemia complementation group (FANC) members FANCI and FANCG revealed a cluster (Fig. 5d, right) that predominantly consists of splice and nonsense variants, with the exception of a single missense FANCI variant (FANCI.356 (E1258K)) that increases γH2AX foci for OLAP and CISP treatments far more strongly than other FANC missense variants. Clustering of all FANC gene variants also identified clusters with one cluster containing most splice and nonsense variants, showing a similar optical feature signature as in the FANCI–FANCG clustering result (Supplementary Fig. 13c). Besides the missense variants of FANCI, two additional missense variants of FANCM (FANCM.14 (A46V)) and FANCL (FANCL.24 (S113N)) were observed with a similar optical feature signature. Variant clusters with notable signatures can also be found in the clustering outcomes of other functionally related genes, such as BRCA2 and PALB2 (Supplementary Fig. 13a) and ATM (Supplementary Fig. 13b).
Moreover, we performed hierarchical clustering on the 273 guides in the library that have an RS2 on-target score ≥ 0.5 (Supplementary Fig. 14). A zoomed-in version of the three top clusters (Supplementary Fig. 15) shows a significant enrichment in RAD51 paralog (RAD51D, RAD51C and XRCC3) variants in the top cluster (P = 1.1 × 10−5, hypergeometric test), featured by downregulation of RAD51 foci across all treatments and upregulation of γH2AX foci in CISP-treated and OLAP-treated cells. We also observed that ATR, BRCA1 and BRCA2 variants clustered together with these RAD51 paralog variants, showing similar patterns of phenotypic changes, suggesting a disrupted homologous recombination response to DNA double-stranded breaks. In addition, strong enrichment of ATM variants was observed by the second top cluster (P = 5.4 × 10−6, hypergeometric test), where γH2AX foci are significantly reduced in ETOP-treated cells and mostly downregulated in CISP-treated and OLAP-treated cells. FANC family gene variants were observed to be enriched in the third top cluster (P = 3.2 × 10−5, hypergeometric test), which features an upregulation of γH2AX foci in OLAP-treated and CISP-treated cells.
Beyond the investigation of phenotype change in each type of foci, we performed co-localization analysis on the six DDR foci (RAD51, BRCA1, RPA2, γH2AX, 53BP1 and RAD18) (Methods) to investigate the foci-to-foci co-localization as a result of perturbations. We performed this analysis on the ionizing radiation dataset, as our main focus was on BRCA1–RAD51 and BRCA1 or RAD51 co-localizations with other foci during the process of homologous recombination. To confirm that the foci co-localize due to biological reasons and not by random chance, we simulated the chance of random co-localization within the nucleus and observed that foci co-localizations are not random (P < 0.0001; Methods and Supplementary Fig. 16a). We then compared the differences in abundance of foci pair co-localizations between G1 phase and S/G2 phase, and we observed an increase in BRCA–RAD51 co-localization in the S/G2 phase as expected (Supplementary Fig. 16b). We also calculated the proportion of individual foci co-localizing with another foci, and, as expected, we observed a high proportion of RAD51–BRCA1 foci co-localization in the S/G2 phase, whereas high proportions were observed between 53BP1–γH2AX foci in both G1 and S/G2 phases (Supplementary Fig. 16c,d). These results highlight the possibility of investigating higher-order interactions between different proteins involved in DDR in future studies.
As a whole, these data support that CRISPRmap not only empowers the analysis of missense VUSs by their correlation with known pathogenic splice or nonsense variants on functionally related genes but also characterizes the drug-specific responses of these gene variants for multiple DDR pathway regulators. The high multiplexed nature of CRISPRmap phenotyping further identifies unique optical signatures of variant clusters that sheds light on the molecular mechanisms of known pathogenic variants as well as their correlated VUSs, making CRISPRmap a potential tool to advance patient-specific precision medicine strategies.
CRISPRmap couples barcode detection with cyclic immunofluorescence in tissue
To evaluate if we could read CRISPRmap barcodes in a tissue context, we performed a pilot study and transduced Cas9− OE19 (human esophageal carcinoma) cells with the aforementioned DDR lentiviral library. After antibiotic selection and expansion, 5 million cells were suspended in a 1:1 mixture of Matrigel and PBS and inoculated into the flanks of a nude mouse. Tumor tissue was harvested after 17 d and flash frozen (Fig. 6a). The CRISPRmap protocol was performed on sectioned tissue with minor modifications (Methods) to yield ample barcode detection throughout the section (Fig. 6c) in recognizable epithelial growth patterns, suggesting that single cells yielded local clonal outgrowth. Subsequent multiplexed immunofluorescence profiling enabled cell and nucleus segmentation based on E-cadherin (Fig. 6e) and DAPI stains, respectively. Approximately 76% of the nuclei in the tissue sections were contained in cell segmentation mask obtained from E-cadherin stains, indicating that about one-fourth of the tumor cells are non-cancer cells. Evaluation of barcode signal for segmented cells revealed that 56% of segmented cells passed our barcode QC metrics (Fig. 6b), and the median number of barcodes detected per cell was 14 (Supplementary Table 4). We expect that the lower number of cells with barcode assignment is due to a variety of reasons, including a lack of antibiotic selective pressure during 17 d of tumor growth, enabling cells to silence the barcode expression. Furthermore, imperfect cell segmentation of the morphologically diverse cancer cells can complicate barcode purity and segment small non-cancer cells by mistake. Analysis revealed that cells with a mask size of less than 250 pixels show a significantly lower count of barcode-assigned amplicons (P = 9.3 × 10−56, two-sided Mann–Whitney test) with a median of 0.0 spots and a mean of 2.2 spots, compared to the rest of the cells with a median of 5.0 spots and a mean of 12.4 spots (Supplementary Table 4). These small cells contribute to 7.1% of the cells that we segmented and analyzed. Another mechanism that leads to poor barcode readout is cell death. We performed an analysis based on the average fluorescence intensity of the cleaved PARP staining under cell nuclei masks and classified 3.6% cells as cPARP+. We observed a significantly lower spot count (P = 4.0 × 10−18, two-sided Mann–Whitney test) with a median of 0.0 spots for the most represented barcode in cPARP+ cells compared to a median of 5.0 spots in cPARP− cells (Supplementary Table 4).
a, Experimental workflow. Cas9− OE19 cells were transduced with the 364-guide DDR library and selected with puromycin for 2 d, before inoculation in the flanks of nude mice. Tumors were harvested after 17 d of growth and processed for CRISPRmap and immunofluorescence imaging. Image was made using BioRender. b, Quantification of proportion of cells segmented on the E-cadherin stain passing the barcode QC criteria (blue; Methods) and proportion of E-cadherin segmented cells that is part of a clonal region (orange; Methods). Data are presented as mean values ± 95% confidence interval (CI). n = 3 technical replicates. c, Visualization of in vivo barcode detection, showing the guide distribution landscape in a tumor section. Decoded barcodes are shown as spots, false colored according to their guide identity. The region highlighted by a white dashed square is zoomed in on e–h. Scale bar, 200 μm. d, Clonality analysis of barcoded cells in a cell-centric manner based on 10-nearest neighbor graphs (Methods). Scale bar, 50 μm. e, Cell (green) and nuclear (blue) boundaries detected by segmentation of E-cadherin and DAPI, respectively. Subcellular resolution of barcode readout. Decoded barcodes are shown as spots, false colored according to their guide identity. f, Iterative immunofluorescence distinguishes cell types and cellular states in vivo. Protein stains of tenascin C (magenta), mouse CD31 (green) and DAPI (blue) are shown. Antibodies are predicted to recognize epitopes from both human and mouse origins unless otherwise specified. g, As in f for vimentin (magenta) and human p21 (green). h, As in f for N-cadherin (magenta) and E-cadherin (green).
In addition, we performed clonality analysis27 of barcoded cells in a cell-centric manner based on 10-nearest neighbor graphs (Methods). Cells with significant enrichment for their barcode in the 10-nearest neighbors, along with the same guide cells in the neighbor graph, were plotted to identify clonal regions (Fig. 6d). This analysis revealed that 36% of E-cadherin segmented cells (or 64% of barcoded cells) are considered part of a clonal region (Fig. 6b). Analysis of interaction patterns between clones17 shows that subcutaneous OE19 tumors have a predominantly clonal distribution (Supplementary Fig. 17a) when compared to other reported tumor types17.
We observed a strong skew of the library of guides detected across the three different tissue sections evaluated (Supplementary Fig. 17b,c). Out of the 364 guides present in the DDR library, we observed 192 guides with at least three cells across the tissue sections evaluated, of which 133 were present in clonal regions. Based on literature, we expect the degree of library skew to be cancer/model dependent; as such, we provide an estimate of the tumor tissue area needed to profile as a function of the number of barcoded cells per guide and assuming the relative frequency in the tissue is that of the one observed in the plasmid library (Supplementary Fig. 17d,e).
Further antibody staining cycles allowed for the visualization of angiogenesis (CD31, endothelial marker; Fig. 6f), extracellular matrix formation (tenascin C; Fig. 6f) around tumor domains as well as a layer of cells expressing vimentin (Fig. 6g) and N-cadherin (Fig. 6h) and transcription factor nuclear translocation in the transplanted cells (p21; Fig. 6g). We observed areas in the tumor tissue without cells and wondered if this is due to cell loss during tissue preparation. We evaluated the loss of nuclei between the first cycle of imaging (barcode readout) and the last round of imaging (antibody stain), and we observed that 95.6% of nuclei can be registered between these rounds (Supplementary Fig. 17g), indicating that we do not observe substantial loss during barcode and antibody readout rounds. Next, we noticed that cells facing the areas without cells, which we termed ‘voids’, tend to be positive for either CD31 or cPARP staining. Voids were annotated (Methods) as vasculature if cells around the void were enriched for CD31 staining and as necrotic areas if enriched for cPARP staining (Supplementary Fig. 17f).
Iterative immunofluorescence and optically resolved transcriptomics approaches have been established for comprehensive profiling of intracellular, cellular, extracellular and signaling mechanisms in the tumor microenvironment. Coupling spatial genomics to CRISPRmap thus enables in vivo CRISPR screens at subcellular resolutions to systematically interrogate how genomic alterations and protein function modify cellular behavior in a tissue context and/or effects upon the cellular microenvironment. Moreover, CRISPRmap is a CRISPR enzyme agnostic barcode readout and, thus, adaptable to the wider CRISPR toolkit, including base editing, gene activation and epigenetic modifications, enhancing the potential to uncover the influence of genes and their regulatory mechanisms on cellular organization and microenvironment.
Discussion
Our study introduces CRISPRmap, a sequencing-free in situ CRISPR barcode readout approach coupled with cyclic immunofluorescence and in situ RNA detection. CRISPRmap expands upon traditional boundaries of optical pooled genetic screening. The AND logic used by CRISPRmap is designed to increase detection specificity when compared to single oligo detection approaches because it requires the adjacent hybridization of both the primer and padlock detection oligos. For approaches that require only a single detection probe that carries the full barcode, and do not require a gap-fill reaction, any non-specific oligo could give rise to an amplicon if the 5′ and 3′ ends of the detection oligo are templated by a random oligonucleotide during the ligation reaction.
Extending the AND logic to the splints further increases specificity when compared to approaches that use primer and padlock oligos but where only the padlock carries barcode information. In CRISPRmap, we distributed the barcode across the primer and padlock detection oligos such that we can identify possible self-ligations of the padlock (only positive for two readout probes) or unallowed combinations of padlock and primer oligos. Collectively the AND requirement for the primer, padlock and two splint oligos is likely to promote specificity. CRISPRmap lacks a reverse transcription or gap-fill reaction typically required for in situ sequencing approaches, which likely contributes to increased efficiency of detection.
Of note, the degree of multiplexed phenotypic readout of our approach can be further expanded for both protein and transcriptomic detection. In this study, although we profiled only 12 transcripts, our RNAmap design is extendable to detect the expression of hundreds to thousands of genes, similar to recent hybridization-based optical transcriptomic approaches18,19,20,28, which have profiled the expression of hundreds to thousands of genes, although such approaches can have practical limits associated with optical crowding and limited dynamic range. Multiplexed immunofluorescence assays, such as IBEX, have achieved concurrent profiling of approximately 60–100 protein targets22. In expanding the antibody panel for large-scale screens, careful consideration should be given to avoid tissue damage during cyclic staining and steric hindrance between antibody panel members. This study enriches understanding of gene functions by enabling systematic examination of spatial phenotypes within perturbed cells—attributes such as morphology and subcellular localization of proteins that are lost in sequencing-based methodologies.
Our approach reduces the costs associated with barcode readout and minimizes reliance on proprietary sequencing reagents. Moreover, CRISPRmap is flexible in the choice of readout dyes, so they can be matched to existing microscopy setups available to researchers, encouraging broad adoption in the community. In addition to cancer cell lines profiling in vitro and in vivo, CRISPRmap is also applicable in primary cells, stem cells, induced pluripotent cells and neurons derived from pluripotent cells, expanding the applicability of optical pooled screens to more challenging cell types in comparison to conventional OPS approaches.
We applied CRISPRmap to investigate the functional consequences of nucleotide variants in genes critical for DDR. Our multimodal profiling of approximately 1 million cells enables a nuanced interrogation of how these variants influence cellular response to DNA damage by expanding the phenotypic profiling from fitness or a single fluorescent reporter to measuring dozens of DDR genes at the proteomic and transcriptomic level with subcellular spatial resolution. This enhanced view of the DDR response empowered us to identify missense VUSs whose DDR response resembles known pathogenic nonsense or splicing variants more closely than most VUSs or unknown missense variants. Notably, our study was carried out in a breast cancer cell line, a disease for which further annotating VUSs for their pathogenic potential is likely to help patients prioritize therapeutic strategies. As such, our approach can provide a framework for annotation of human variants in a treatment-specific manner and can help prioritize therapeutic strategies.
Recently, an elegant use of triplet combinations of linear epitopes enabled antibody-based identification of CRISPR guide expression of dozens of different guide-expressing cancer cell populations within a tumor tissue at single-cell resolution and tissue scale17. RNA-based barcoding offers an opportunity to increase library complexity to genome scale29 but, to date, has not been established in a tissue context. We expect that CRISPRmap can be scaled up to accommodate larger screening library sizes. The set of 54 primer and 54 padlock oligos used in this study enables up to 2,916 barcodes, but scaling up to 231 primer and padlock probes would encode for 53,361 barcodes and, thus, enable genome-wide screening with two sgRNAs per protein-coding gene, which could be read out using 44 20mer readout probes across 15 barcode readout cycles. Although genome-wide screens are feasible in the in vitro setting, in vivo studies should carefully consider how many different perturbations can be profiled in a given mouse model and how much tissue should be profiled to establish phenotypic significance. As reported in this study, CRISPRmap can detect ample barcode signal in a tissue context with subcellular resolution, which enables the interrogation of cells with their neighbors and their surrounding extracellular milieu as a function of precise genome editing. Another opportunity for RNA-based barcoding is to start exploring the effects of combinatorial gene perturbations, enabled by the spatial resolution of the barcode that offers the capability to detect more than one barcode expressed per cell. This capability could be helpful for screens to prioritize combinatorial targets and inform therapy modalities that go beyond single-therapy strategies.
However, our study is not without limitations. RNA-based barcoding approaches are reliant on the stability of RNA molecules, which can be challenging in a tissue context due to the presence of RNAse enzymes. A possible resolution for this reliance is to transcribe barcode-carrying RNA in vitro by T7 polymerases post hoc, as was recently reported for in vitro cells30. This approach enables deep phenotypic profiling of tissue before in vitro generation of barcode-carrying RNA molecules that can subsequently be detected by CRISPRmap. Moreover, although our RNAmap profiling approach has demonstrated good specificity, the detection efficiency is lower than traditional single-molecule fluorescence in situ hybridization (smFISH) approaches. RNAmap allows for strong signal amplification, which enables high-throughput imaging (×20 objective and short exposure times), thus enabling visualizing millions of cells needed for large-scale screening. For any study at hand, it would be of interest to evaluate the balance of deeper transcriptomic profiling enabled by FISH-based approaches and throughput and scale of the screen. We expect CRISPRmap to be fully compatible with FISH-based transcriptomic profiling. These constraints highlight the need for continual refinement of optical screening techniques and computational analysis methods.
Future directions could focus on expanding the versatility of CRISPRmap to include a broader range of CRISPR modalities, cell types and tissue environments. Studies can also delve deeper into the impact of genetic perturbations on tissue architecture and the interplay between cells in complex microenvironments. We envision that CRISPRmap will pave the way for high-throughput investigations of gene function in diverse biological contexts, from developmental biology to the study of disease pathogenesis.
In conclusion, CRISPRmap offers a new lens through which the intricate tapestry of gene function within and across cells can be examined. Our findings herald a shift toward more spatially and temporally resolved studies of gene function, especially in tissue environments, potentially illuminating new paths in precision medicine and the quest to understand the underpinnings of complex biological systems.
Methods
Cell lines and cell culture
HEK293FT cells (Thermo Fisher Scientific, R70007) were cultured in DMEM (Gibco, 11965092) supplemented with heat-inactivated 10% FBS (American Type Culture Collection (ATCC), 30–2020) and 100 U ml−1 penicillin–streptomycin (Thermo Fisher Scientific, 15140163). MCF7-BE3 cells were cultured in the same medium supplemented with 2 μg ml−1 blasticidin (Thermo Fisher Scientific, A1113903). HT1080–Cas9 AAVS1 (GeneCopoeia, SL512) cells were cultured in the same medium supplemented with 200 μg ml−1 Hygromycin B Gold (Invivogen, ant-hg-2). OE19–BFP cells were cultured in RPMI 1640 medium (ATCC, 30–2001) supplemented with 10% heat–inactivated FBS, 100 U ml−1 penicillin–streptomycin, 1× GlutaMAX supplement (Thermo Fisher Scientific, 35050079) and 2 μg ml−1 blasticidin. IMR-90 (ATCC, CCL-186) cells were cultured in EMEM (ATCC, 30–2003) supplemented with heat-inactivated 10% FBS and 100 U ml−1 penicillin–streptomycin. Rockefeller University Embryonic Stem Cell Line 2 (RUES2, passages 24–32) were maintained on mouse embryonic fibroblasts (MEFs) (Thermo Fisher Scientific, A34180) and plated at 22,500 cells per cm2. Cells were cultured in hESC maintenance media (DMEM/F12 (Thermo Fisher Scientific, 11320033), 20% knockout serum (STEMCELL Technologies), 0.2% Primocin (InvivoGen, ant-pm-05), 0.1 mM β-mercaptoethanol (Sigma-Aldrich, M6250), 20 ng ml−1 FGF2 (R&D Systems, 233-FB) and 1% GlutaMAX). The medium was changed daily. hESCs were passaged every 3–4 d with Accutase (Innovative Cell Technologies, AT-104), washed and replated at a dilution of 1:24. Cultures were maintained in a humidified 5% CO2 atmosphere at 37 °C. Lines are karyotyped and verified for mycoplasma contamination using PCR every 6 months. hESCs were infected with the virus for 24 h in hESC medium supplemented with polybrene and puromycin selected. Before analysis, MEFs were depleted by passaging 5–7 × 105 hESCs onto Matrigel (Corning, 354277, dilution 1:15)-coated flat glass-bottom 96-well plates. Cells were maintained in hESC medium in a humidified 5% CO2 atmosphere at 37 °C. For human iPSC and iMN experiments, we used reference Wt line KOLF2.1J (ref. 31) (a gift from Christopher Ricupero). iPCSs were maintained on Matrigel (Corning, 354277, dilution 1:100)-coated plates in mTeSR Plus media (STEMCELL Technologies, 100–0276), supplemented with Y-27632 (ROCKi, 10 µM, Selleckchem, S1049) during thawing, passaging and viral transduction. Passaging was performed using Accutase (Thermo Fisher Scientific, A1110501). iPSCs were transduced with the CRISPRmap library during passaging by adding viral supernatant to polybrene (8 μg ml−1, Sigma-Aldrich, TR-1003-G)-supplemented media at various dilutions. Forty-eight hours later, transduced cells were selected with puromycin (1 µg ml−1, Thermo Fisher Scientific, A1113802) for 3 d. For optical barcode detection in iPSCs, iPSCs were dissociated into single cells and plated on polyethylenimine (PEI, Sigma-Aldrich, 408719)-coated 96-well plates for imaging in mTeSR Plus with ROCKi at 10,000 cells per well. ROCKi was maintained before fixing, preventing tight colony formation to simplify cell segmentation during image analysis. For coating, 96-well plates were incubated at 37 °C overnight with PEI (250 µg ml−1, 50 µl per well) and then washed at least three times with 200 µl per well PBS.
iPSC to iMN differentiation was carried out as previously described32,33. In brief, on day 0, iPSCs were dissociated to single cells with Accutase and resuspended in N2B27 differentiation media (1:1 Advanced DMEM/F12 and Neurobasal media (Life Technologies, 12634010 and 21103049)), GlutaMAX (1%, 35050061), β-mercaptoethanol (0.1%, Sigma-Aldrich), N-2 (1%, Thermo Fisher Scientific, 17502048), B-27 (2%, Thermo Fisher Scientific, 17504044) and ascorbic acid (10 μM, Sigma-Aldrich, A4403), supplemented with ROCKi (10 µM), FGF2 (10 ng ml−1, PeproTech, PHG0263), CHIR99021 (CHIR, 3 µM, Tocris, 4423), SB 431542 hydrate (SB, 20 μM, Sigma-Aldrich, S4317) and LDN193189 (LDN, 100 nM, Stemgent, 04-0074) at a density of 50,000 cells per milliliter on ultra-low adhesion dishes to promote embryoid body (EB) formation. On day 2, media were replaced, supplemented with CHIR (3 µM), SB (20 µM), LDN (100 nm), all-trans retinoic acid (RA, 100 nM, Sigma-Aldrich, R2625) and smoothened agonist (SAG, 500 nM, Millipore, 566660). On day 4, media were replaced and supplemented as on day 2. On day 7, media were replaced and supplemented with RA (100 nM), SAG (500 nM) and BDNF (10 ng ml−1, PeproTech, 450-02). On day 9, media were replaced and supplemented with RA (100 nM), SAG (500 nM), BDNF (10 ng ml−1) and DAPT (10 µM, Selleckchem, S2215). On day 11, media were replaced and supplemented as on day 9. On day 14, media were replaced and supplemented with RA (100 nM), SAG (500 nM), BDNF (10 ng ml−1), DAPT (10 µM) and GDNF (10 ng ml−1, R&D Systems, 212-GD-050). On day 16, EBs were dissociated to single cells by trituration with 0.05% trypsin (Life Technologies, 25300054). Dissociated MNs were resuspended in hMN maintenance media (Neurobasal media, GlutaMAX (1%), NEAA (1%, Life Technologies, 11140050), β-mercaptoethanol (0.1%), N-2 (1%), B-27 (2%), ascorbic acid (10 μM), BDNF (10 ng ml−1), GDNF (10 ng ml−1), CNTF (10 ng ml−1, PeproTech, 257-NT-050), IGF-1 (10 ng ml−1, PeproTech, 291-G1), RA (1 µM) and adarotene (1 µM, MedChemExpress, HY-14808)) and plated on PEI-coated 96-well plates for imaging at 10,000 cells per well.
After CRISPRmap amplicon generation, cell type validation was performed on iPSCs and iMNs by immunostaining using anti-SOX2 (iPSC marker, 1:200, Thermo Fisher Scientific, 14-9811-82), anti-OCT4 (iPSC marker, 1:200, Cell Signaling Technology, 2840) and anti-NeuN (iMN marker, 1:200, Millipore Sigma, MAB377) antibodies. Likewise, cell type validation was performed on hESCs using anti-SOX2 (hESC marker, 1:200), anti-OCT4 (hESC marker, 1:200) and anti-Nanog (hESC marker, 1:200, Cell Signaling Technology, 4893) antibodies.
Library design and cloning
GFP-targeting guides in the GFP-targeting CRISPRmap knockout screen library (referred to as GFP-pilot) were designed with CRISPick34 by selecting the top five recommended candidates in the CRISPRko mode that target the copGFP sequence. The library also contains five NTC guides that lack targets in the human genome. Each guide was combined with a universal scaffold sequence and a pair of guide-specific CRISPRmap barcode sequences. Universal 5′ and 3′ homology sequences were then added to facilitate NEB HiFi assembly into the expression vector. Full-length GFP-pilot library sequences are shown in Supplementary Table 5. Base editing guides in the DDR screen library (referred to as DDR364) were selected from the base editing screens as previously described7. The library contains 162 missense guides and 50 nonsense guides with a single C base in the editing window (4th to 8th base in the guide targeting sequence), 80 splice-donor or splice-acceptor (referred to as splice) guides, 35 guides targeting the AAVS1 safe-harbor site and 37 NTC guides that have minimal targets in the human genome. All the selected missense, nonsense and splice guides have false discovery rate (FDR) < 0.05 in at least one treatment in the previous screen7. Similarly, each guide was combined with the scaffold, CRISPRmap barcode and homology sequences. Sequences are shown in Supplementary Table 6. Both libraries were ordered as synthesized oligo pools (Integrated DNA Technologies) and PCR amplified with Q5 DNA polymerase (New England Biolabs, M0492) using an optimized two-round amplification strategy to minimize barcode–sgRNA recombination35. In brief, oligo pools were diluted in ultrapure water (Thermo Fisher Scientific, 10-977-023); 1 pg of total DNA was added to each 50-μl Q5 reaction mix to perform the first-round amplification of 15 PCR cycles; and 0.5 μl of PCR product from each 50-μl first-round reaction was then added to each 50-μl Q5 reaction mix for the second-round amplification of 10 cycles. Final PCR product was purified with DNA Clean & Concentrator (Zymo Research, D4013). The primer pairs CRISPRmap-F and CRISPRmap-R in Supplementary Table 7 were used in both rounds. Amplified oligo pools were cloned into a modified CROPseq-puro-v2 (Addgene, 127458) vector that removed the original scaffold sequence (referred to as CRISPRmap-CROPseq) using NEBuilder HiFi DNA Assembly (New England Biolabs, E2621). Next, we electroporated into MegaX DH10B electrocompetent cells (Thermo Fisher Scientific, C640003). An average number of 300 colonies per guide was maintained to preserve the relative abundance of guides in the library. Bacterial colonies were scraped and pooled for plasmid extraction (Zymo Research, D4212).
Lentivirus production and titer determination
293FT cells were seeded into six-well tissue culture–treated plates at a density of 100,000 cells per cm2. After 24 hours, cells were transfected with pMD2.G (Addgene, 12259), psPAX2 (Addgene, 12260) and CRISPRmap library plasmid (2:3:4 ratio by mass) using Lipofectamine 3000 (Thermo Fisher Scientific, L3000001) in Opti-MEM (Thermo Fisher Scientific, 31-985-070), supplemented with 5% FBS. Media were exchanged after 6 h and supplemented with 1.5 mM caffeine (Sigma-Aldrich, C0750) to increase viral titer. Viral supernatant was harvested at 24 h and 48 h after transfection, filtered through 0.45-μm cellulose acetate filters (Corning, 431220) and stored in a −80 °C freezer in aliquots.
Lentiviral titer was determined by the colony formation assay to control the MOI in downstream studies. In brief, 10-fold serial dilutions of the lentivirus stock were prepared in complete DMEM containing 8 μg ml−1 polybrene. In total, 10,000 cells were seeded into each well of a six-well plate. A total volume of 1 ml of diluted lentivirus was added to each well for 48 h. Cells were then cultured in complete DMEM supplemented with appropriate antibiotics for 14 d, and media were changed every 3 d. Cells were fixed and stained with 0.1% crystal violet (Sigma-Aldrich, V5265) for 10 min at room temperature and washed three times with PBS. Colonies on each well were counted, and the transduction units per milliliter (TU/ml) was calculated as follows: TU/ml = number of colonies / total volume in the well (ml) × dilution factor.
Fluorescence microscopy
All imaging datasets were acquired using a confocal spinning disk microscope (Andor Dragonfly) coupled to a Nikon Ti-2 inverted epifluorescence microscope with automated stage control, Nikon Perfect Focus System and a Zyla PLUS 4.2-megapixel USB3 camera. Illumination was done with 100 mW 405 nm, 50 mW 488 nm, 50 mW 561 nm, 140 mW 640 nm and 100 mW 785 nm solid-state lasers. All hardware was controlled using Andor Fusion software. Lasers, laser powers, exposure times, objectives and experiment-specific acquisition parameters are summarized in Supplementary Tables 5 and 6. Images were acquired with four z-slices at 1.5-μm intervals for the cultured cells and with six z-slices at 1.5-μm intervals for the tissue sections unless otherwise specified.
Oligonucleotide fluorophore conjugation
In each 10-μl reaction, 2 μl of 0.5 mM 5′ amine-modified DNA probes (Integrated DNA Technologies) was mixed with 1 μl of 10 mM ATTO488-NHS ester (ATTO-TEC AD, 488-31), ATTO 643-NHS ester (ATTO-TEC AD, 643-31) or CF568 succinimidyl ester (Sigma-Aldrich, SCJ4600027) in 1× BBS (Thermo Fisher Scientific, 28384), pH 8.5, and incubated at room temperature for 4 h. Fluorophore-conjugated DNA probes were purified with Oligo Clean & Concentrator (Zymo Research, D4060) and diluted to 1 μM in ultrapure water, aliquoted and stored at −20 °C. Oligonucleotide sequences and fluorophores used in the GFP-targeting screen are listed in Supplementary Table 5, and the base editing screens and in vivo CRISPRmap barcode readout are listed in Supplementary Table 6.
Antibody fluorophore conjugation
In each conjugation reaction, 5 μg of antibody in PBS (BSA-free) is mixed with 1 μl of 0.33 mM CF750 Dye SE/TFP esters (Biotium, 92142), Alexa Fluor 647 NHS Ester (Thermo Fisher Scientific, A20006), Alexa Fluor 555 NHS Ester (Thermo Fisher Scientific, A20009) or Alexa Fluor 488 NHS Ester (Thermo Fisher Scientific, A20000) in DMSO and incubated at room temperature for 16 h. Fluorophore-conjugated antibodies were then purified with a 30-kDa Amicon Ultra-0.5 Centrifugal Filter Unit (Millipore Sigma, UFC5030BK) Antibodies used in the base editing screen and in vivo barcode readout are listed in Supplementary Table 6.
CRISPRmap optical pooled CRISPR knockout screen
HT1080–Cas9 AAVS1 cells were seeded into six-well tissue culture–treated plates at a density of 50,000 cells per cm2. After 24 h, cells were transduced with the GFP-pilot lentiviral supernatant supplemented with 8 μg ml−1 polybrene at MOI ~ 0.1. At 48 h after infection, viral supernatant was removed, and cells were treated with media containing 2 μg ml−1 puromycin for 48 h and seeded onto 96-well glass-bottom plates (Cellvis, P96-1.5H-N) at 10,000 cells per well as the original seeding density. Cells were seeded at 4,000 cells per well as the sparse density to avoid extensive overlapping among cells. A total reaction volume of 50 μl was used in the following steps unless otherwise specified. After 24 h, cells were fixed in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences, 15710-S) in PBS (Gibco, 10010049) for 10 min at room temperature, followed by two rinses in PBS. Cells were then incubated in 0.1 mg ml−1 wheat germ agglutinin (WGA) CF770 conjugate (Biotium, 29059) and 0.5 μg ml−1 DAPI (Abcam, ab285390) in PBS for 30 min at room temperature and imaged in PBS for membrane, GFP and nuclei signal using the microscope configuration described above. After phenotype imaging, cells were permeabilized with 0.2% Triton X-100 (Sigma-Aldrich, T8787) in PBS for 10 min at room temperature, followed by two rinses in PBS. The permeabilization conditions are to be determined for each new cell type, as it is one of the parameters that determines barcode detection efficiency. For primer and padlock oligo hybridization, cells in each well were incubated in the hybridization mix (GFP-pilot CRISPRmap padlock and primer mix (see Supplementary Table 5 for sequences; each oligo in the mix has a final concentration of 10 nM), 1 mg ml−1 yeast tRNA (Invitrogen, 15401011) and 2× SSC, 20% formamide (v/v) in ultrapure water) for 16 h at 40 °C in a HybEZ oven (ACD, PN 321720). After hybridization, cells were first rinsed three times with the hybridization wash buffer (2× SSC, 20% formamide (v/v) in ultrapure water) and then washed three times for 5 min at 40 °C. Cells were then incubated in splint mix (10 nM CRISPRmap GFP-pilot splint mix (see Supplementary Table 5 for sequences; each splint oligo in the mix has a final concentration of 10 nM), 0.1% yeast tRNA, 2× SSC and 15% formamide in ultrapure water) for 30 min at 37 °C in a HybEZ oven, rinsed twice with the formamide wash buffer (2× SSC, 15% (v/v) formamide in ultrapure water) and incubated in 2× SSC in ultrapure water for 15 min at room temperature. For T4 DNA ligation, cells were incubated in ligation mix (1× T4 ligase buffer, 1% (v/v) T4 DNA ligase (Enzymatics, L6030-HC-L) in ultrapure water) for 2 h at 16 °C and then for 1 h at 25 °C in a HybEZ oven, followed by two rinses in PBS. For RCA, cells were incubated in RCA mix (1× QualiPhi buffer, 2% (v/v) QualiPhi DNA Polymerase (4basebio, 510100), dNTP mix (0.25 mM each; Thermo Fisher Scientific, R1122), and 0.02 mM 5-(3-aminoallyl)-dUTP (Thermo Fisher Scientific, AM8439) in ultrapure water) for 6 h at 30 °C and then the RCA mix was removed and the cells were immediately fixed with 4% PFA in PBS for 10 min at room temperature, followed by three PBS washes. For readout probe hybridization, cells in each well were incubated in readout probe mix (10 nM of each readout probe (see readout probe sequences for each hybridization rounds in Supplementary Table 5), 2× SSC, 15% formamide in ultrapure water) for 30 min at 37 °C in a HybEZ oven. Cells were then imaged in the imaging buffer (0.5 μg ml−1 DAPI, 10 μg ml−1 Fungin (InvivoGen, ant-fn-1) in PBS) using the microscope configuration described above. After imaging, the cells were incubated in the stripping buffer (2× SSC, 50% formamide (v/v) in ultrapure water) for 20 min at 40 °C in a HybEZ oven and then rinsed once with formamide wash buffer. A total of four readout hybridization rounds were performed to decode all the CRISPRmap barcodes in the GFP-pilot library. The same CRISPRmap assay and barcode detection protocol was applied to IMR-90 cells, iPSCs, iMNs and hESCs.
To quantify the sensitivity, specificity and precision of the assay, average fluorescence intensity of GFP under nuclei masks was quantified, and a threshold was determined based on the GFP intensity distribution of the cell population to classify cells into GFP+ and GFP− categories. Standard CRISPRmap QC was performed for each cell to determine the guide identity. Specifically, we quantified the spot count for the most representing guide-reporting barcode (max_spot) and the second most representing guide-reporting barcode (second_max_spot) in each cell, and a purity score was calculated by: Purity = max_spot / (max_spot + second_max_spot). Guide identity is only assigned to a cell (that is, a cell that passed QC) when the max_spot ≥ 3 and Purity ≥ 0.66. A relaxed QC metric (max_spot ≥ 2 and Purity ≥ 0) was applied to enable a side-by-side comparison between CRISPRmap and conventional OPS. The guide identity reported by the most representing barcode is assigned to a cell that passed QC. Ratio of cells that passed QC was calculated as the ratio between the number of cells that passed QC and the total number of cells profiled. To calculate sensitivity, specificity and precision, we define a true positive (TP) as a GFP− cell assigned with one of the GFP− targeting guides in the GFP-pilot library; a true negative (TN) is defined as a GFP+ cell assigned with one of the non-targeting guides; a false positive (FP) is defined as a GFP+ cell assigned with one of the GFP-targeting guides; and a false negative (FN) is defined as a GFP− cell assigned with one of the non-targeting guides. Specificity = TN / (TN + FP); Sensitivity = TP / (TP + FN); Precision = TP / (TP + FP). QC metrics from loose to tight were applied in Supplementary Fig. 1c–e.
CRISPRmap barcode readout optimization and quantification
To optimize and quantify the CRISPRmap barcode readout, we created two HT1080 cell lines with fluorescent protein (FP) expression tethered to the CRISPRmap barcode reporting on the FP identity: one cell line expresses GFP, an NTC guide (NT_GFP) and the CRISPRmap padlock and primer hybridization sequence for Padlock003 and Primer003 (GFP_Barcode); the other cell line expresses mTurquoise2, an NTC guide (NT_mTurquoise2) and the CRISPRmap padlock and primer hybridization sequence for Padlock004 and Primer004 (mTurquoise2_Barcode). The sequences are listed in Supplementary Table 5. FPs were introduced to the CRISPRmap-CROPseq vector by replacing the puromycin resistance gene. The sgRNA–barcode (NT_GFP + GFP_Barcode, NT_mTurquiose2 + mTurquoise2_Barcode) sequences were ordered as synthesized double-strand DNA fragments (Integrated DNA Technologies) and cloned onto CRISPRmap-CROPseq vector replaced with GFP and mTurquoise2, respectively. Plasmid-EZ sequencing (Azenta Life Sciences) was performed to confirm the sgRNA–barcode combination matches with the FP expressed, before lentiviral packaging and infection on the HT1080 cells. The two cell lines were sorted by flow cytometry, mixed at 1:1 ratio and seeded onto 96-well glass-bottom plates for genotype–phenotype mapping. Cells were fixed in 4% PFA in PBS for 10 min at room temperature and then incubated with DRAQ5 fluorescent probes (0.05 mM, Thermo Fisher Scientific, 62251) and WGA-CF770 (10 µg ml−1) for 20 min at room temperature for nucleus and membrane segmentation, respectively. WGA-CF770, DRAQ5, GFP and mTurqoise2 fluorescence signals were imaged with 730-nm, 640-nm, 488-nm and 405-nm lasers, respectively. Cell permeabilization, amplicon generation, barcode readout and image analysis were performed, as described in the CRISPRmap pooled CRISPR knockout screen for the GFP-pilot library. Average fluorescence of GFP and mTurquiose2 signals under nuclei masks was quantified to classify cells into GFP+ and mTurquoise2+ categories, and the FP identity in each cell was matched to the detected barcodes to evaluate the sensitivity, specificity and precision of the assay (Supplementary Fig. 1c–e and Supplementary Table 2).
To quantify the CRSPRmap barcode readout for potential double-transduced cells, we performed lentiviral infection of the GFP-pilot library at different MOI of 0.9, 0.3, 0.1 and 0.03 on HT1080 cells. Cells were puromycin selected, seeded and profiled for barcode expression, as described above. To identify potential double-transduced cells, we first performed standard QC to identify cells with unique barcodes and then classified the remaining cells to be ‘double’ if the spot count for the most representing guide (max_spot) was ≥3 and the second-most representing spot (second_max_spot) was ≥2. The expected ratio of double-transduced cells after antibiotic selection at a given MOI was calculated by Poisson distribution after removing the proportion of cells with zero infection event. The ratio of double-transduced cells detected optically and the expected ratio at each MOI are shown in Supplementary Fig. 1l and Supplementary Table 2.
CRISPRmap multimodal optical pooled base editing screen
MCF7-BE3 cells were transduced with the DDR364 library in the same manner as in the GFP-targeting knockout screen with several modifications to accommodate the multiplexed immunofluorescence and RNAmap. Specifically, after puromycin selection, cells were seeded onto six-well glass-bottom plates (Cellvis, P6-1.5H-N) at a density of 50,000 cells per cm2. For the DDR364-irradiation screening, after 48 h, cells were exposed to 10 Gy of ionizing radiation using the Gammacell 40 cesium source irradiator and fixed at 6 h after irradiation. For the DNA-damaging agents (DDR364-chemo) screening, cells were treated with 100 nM CPT (Sigma-Aldrich, C9911), 1 μM OLAP (Selleck Chemicals, S1060), 1 μM CISP (Sigma-Aldrich, P4394), 1 μM ETOP (Sigma-Aldrich, E1383) or untreated and fixed at 24 h after treatment. After fixation, cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min on ice. Cells in each well were incubated in 1 ml of reaction mix or buffers in all steps unless otherwise specified. After permeabilization, cells were incubated in the antibody mix (2 μg ml−1 rat anti-CD326 (BioLegend, 312502), 1 μg ml−1 rabbit anti-RAD51-AF647 (BioAcademia, 70-012), 2 μg ml−1 mouse anti-BRCA1-AF555 (Santa Cruz Biotechnology, sc-6954), 0.5 μg ml−1 rabbit anti-RPA2-AF488 (Bethyl Laboratories, A300-244A) in PBS) for 1 h at room temperature and then rinsed twice with PBS. Cells were incubated in 10 μg ml−1 goat anti-rat-IgG secondary antibody (Thermo Fisher Scientific, SA5-10023) for 30 min at room temperature and rinsed twice with PBS. Cells were fixed in 4% PFA in PBS for 10 min at room temperature to crosslink the antibodies to the cells, followed by two PBS rinses. Cells were then processed with padlock and primer probe hybridization, splint hybridization, ligation and RCA as described above, with the minor difference that 3 nM of each CRISPRmap padlock and primer probes and 3 nM of each RNAmap padlock and primer probes were used in the hybridization mix. Probe sequences are listed in Supplementary Table 6. After RCA and fixation, cells were first imaged in the imaging buffer for membrane, nuclei and nuclear foci signal using the microscope configuration described in Supplementary Table 6. After imaging, the antibody signal was bleached with 1 mg ml−1 lithium borohydride (Sigma-Aldrich, 222356) and rinsed twice with PBS, before the incubation of the next round of antibodies. For both the DDR364-irradiation and the DDR364-chemo screening, a total of four antibody incubation–bleaching rounds were performed. After the last round of bleaching, eight rounds of RNAmap readout probe hybridization-stripping rounds were performed, followed by eight rounds of CRISPRmap readout probe hybridization-stripping rounds. Each round was imaged using the microscope configuration described above. Readout probe sequences and conjugated fluorophores are listed in Supplementary Table 6. For the DDR364-irradiation screening, cells were incubated in Vector TrueVIEW Autofluorescence Quenching reagent (Vector Laboratories, SP-8400-15) for 5 min at room temperature to reduce autofluorescence, followed by three rinses in PBS before imaging each CRISPRmap readout round in high DAPI imaging buffer (2.5 μg ml−1 DAPI, 10 μg ml−1 Fungin in PBS).
In vivo CRISPRmap barcode readout and multimodal phenotyping
After lentiviral transduction with pLV-EF1a-TagBFP2 on OE19 cells, fluorescence-activated cell sorting (Sony, MA900) was performed to obtain a BFP-expressing OE19 population. BFP-expressing OE19 (referred to as OE19–BFP) cells were lentiviral transduced with the DDR364 library and puromycin selected as described above and then expanded for 4 d in puromycin-free media. We suspended 5 × 106 cells in a 1:1 mixture of Matrigel and PBS and inoculated the mixture into the flanks of nude mice (JAX, strain no. 002019). Mice were housed with a constant temperature of 21–24 °C, 45–65% humidity and a 12-h light/dark cycle. After 17 d, tumors were harvested and fresh frozen in OCT on dry ice and stored at −80 °C. Frozen tumor samples were sectioned using a cryostat microtome (Leica, CM1510S) at −20 °C into 10-μm-thick sections and deposited onto 12-well glass-bottom plates (Cellvis, P12-1.5H-N) coated with 0.1 mg ml−1 poly-d-lysine (Sigma-Aldrich, A-003-E). CRISPRmap barcode readout and antibody staining were performed as described above with minor modifications. Specifically, 400 μl of reaction mix and buffers was added to each well to fully cover the tissue section. Tissue sections were fixed with 4% PFA in PBS for 15 min at room temperature and permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature. Then, 30 nM of each CRISPRmap padlock and primer oligos was used in the hybridization mix. The same set of CRISPRmap padlock and primer probes, splints and readout probes was used as in the DDR364-irradiation screening. Eight CRISPRmap readout cycles were performed before antibody staining and bleaching cycles. The same readout probes were used as in the base editing screens.
Conventional OPS
Conventional OPS on cultured cells (HT1080 cells, IMR-90 cells, iPSCs, hESCs and MNs) was performed in accordance with the published protocol3,4. In brief, cells were fixed and permeabilized in the same conditions with cells undergoing the CRISPRmap protocol for side-by-side comparisons. Specifically, cells were fixed in 4% PFA in PBS for 10 min at room temperature and permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. Reverse transcription mix (1× RevertAid RT buffer, 250 μM dNTPs, 0.2 mg ml−1 BSA (New England Biolabs, B9000S), 1 μM RT primer (/5AmMC12/A + CT + CG + GT + GC + CA + CT + TTTTCAA, Integrated DNA Technologies), 0.8 U μl−1 Ribolock RNase inhibitor (Thermo Fisher Scientific, EO0384) and 4.8 U μl−1 RevertAid H minus reverse transcriptase (Thermo Fisher Scientific, EP0452)) was added to the cells and incubated for 16 h at 37 °C. Cells were washed five times with PBS-T and fixed with 3% PFA and 0.1% glutaraldehyde in PBS for 30 min at room temperature and then washed with PBS-T five times. Cells were incubated with the gap-fill reaction mix (1× Ampligase buffer, 0.4 U μl−1 RNase H (Enzymatics, Y9220L), 0.2 mg ml−1 BSA, 100 nM padlock probe (/5Phos/GTTTCAGAGCTATGCTCTCCTGTTCGCCAAATTCTACCCACCACCCACTCTCCAaaggacgaaaCACC, Integrated DNA Technologies), 0.02 U μl−1 TaqIT polymerase (Enzymatics, P7620L), 0.5 U μl−1 Ampligase (Lucigen, A3210K) and 50 nM dNTPs) for 5 min at 37 °C and 90 min at 45 °C, washed twice with PBS-T and then incubated with the RCA mix (1× Phi29 buffer, 250 μM dNTPs, 0.2 mg ml−1 BSA, 5% glycerol, 1 U μl−1 Phi29 DNA polymerase (Thermo Fisher Scientific, EP0091)) at 30 °C for 16 h. For in situ sequencing, 1 μM sequencing by synthesis primer (GCCAAATTCTACCCACCACCCACTCTCCAaaggacgaaaCACC, Integrated DNA Technologies) in 2× SSC was added to the cells for 30 min at room temperature. Incorporation mix (Illumina, MS-103-1003, MiSeq reagent 1) was added to the cells for 5 min at 60 °C, and the cells were rinsed five times in PR2 and washed by five cycles of 5 min, 60 °C washes. Cells were imaged using iIllumination of 100 mW 405 nm (DAPI), 50 mW 488 nm (G base), 50 mW 561 nm (C base) and 140 mW 640 nm (A base) lasers. Cells were then incubated in the cleavage mix (Illumina, MS-103-1003, MiSeq reagent 4) at 60 °C for 6 min, followed by three rinses with PR2, one wash with PR2 at 60 °C for 1 min and three rinses with PR2 again, before entering the next incorporation step. Four bases were sequenced to distinguish the guide sequences in the GFP-pilot library. Sensitivity, specificity and precision were calculated based on the barcode identity and GFP expression level in each cell, as described in the CRISPRmap pooled CRISPR knockout screen for the GFP-pilot library.
Variant annotation
The sgRNA category of each guide was annotated as previously described7. We grouped the splice-donor and splice-acceptor categories into a ‘splice’ category. All AAVS1-targeting and non-targeting guides are annotated as a ‘control’ category. The ClinVar category was determined by querying each guide in the ClinVar database (version 2023-12-15; https://www.ncbi.nlm.nih.gov/clinvar/; RRID: SCR_006169). Nonsense and missense variants were queried based on the specific amino acid change, whereas splice variants were queried based on the nucleotide change outcomes in the editing window (base C in the 4th to 8th bases in the sgRNA targeting sequences). Note that if multiple C bases exist in the editing window, a splice guide can render other mutational outcomes, such as missense or intron variants. These mutational outcomes were not counted in the annotation of splice variants but listed as ‘Less deleterious variants’ in Supplementary Table 6. The determining criteria of the ClinVar category were established as previously described7. In brief, three categories were assigned to non-control guides: (1) benign/likely-benign (B/LB); (2) VUSs; and (3) P/LP. The VUS category also includes variants with conflicting interpretations. If a variant was not documented in the ClinVar database, it was listed as ‘unknown’.
Library QC and quantification by NGS
For oligo pool quantification, the first-round amplification product in the library cloning step was collected, and 0.5 μl of 50-μl PCR product was added to each 50-μl Q5 reaction mix for the second-round amplification of 10 cycles using the primer pairs CRISPRmap-F-ad and CRISPRmap-R-ad in Supplementary Table 7. We amplified 10 pg of plasmid extraction product from the library cloning step with the same two-round strategy as the oligo pool quantification for plasmid-level quantification. Genomic DNA of the cells transduced with the sgRNA library was extracted with Genomic DNA Clean & Concentrator (Zymo Research, D4010). We amplified 100 ng of genomic DNA with the same two-round strategy. We had 5 ng of the final PCR product sequenced with NGS (Azenta Life Sciences, Amplicon-EZ). sgRNA sequences in the library were aligned to the NGS reads to quantify the relative abundance of each guide in the library, and the padlock and primer hybridization sequences (barcodes) were aligned to each NGS read containing a valid sgRNA sequence to evaluate the barcode–sgRNA recombination rate. Each read with a valid sgRNA sequence was classified into ‘matched’ (sgRNA–barcode combination matched the codebook), ‘switched’ (sgRNA–barcode combination does not match the codebook), ‘loss of BC’ (no valid padlock or primer sequences detected) or ‘unallowed BC’ (unallowed padlock and primer combination detected) category. The results are shown in Supplementary Fig. 1j and Supplementary Table 2.
Base editing screen hits validation
Individual sgRNAs with the same guide and scaffold sequences as used in the base editing screen were ordered as synthesized double-strand DNA fragments (Integrated DNA Technologies) and cloned onto the CRISPRmap-CROPseq vector. As described in the base editing screening, cells transduced with individual sgRNAs were selected for 2 d in puromycin and cultured for 2 d before ionizing radiation. Six hours after irradiation, cells on the glass-bottom plates were fixed for immunostaining of the same panel of nuclear foci imaged in the screen. Cells on tissue culture plates were harvested for immunoblotting. Genomic DNA was extracted from the untreated cells with QuickExtract DNA Extraction Solution (Lucigen, QE09050) at the same timepoint for evaluating base editing efficiency of the individually transduced guides. PCR amplification was performed on the genomic locus of the intended base edit (primer sequences listed in Supplementary Table 7) using Q5 DNA polymerase (New England Biolabs, M0492), followed by Sanger sequencing (Azenta Life Sciences). ICE analysis (Synthego Performance Analysis, ICE Analysis, 2019) was performed on the Sanger sequencing results to quantify the in-window and out-of-window editing efficiency (Supplementary Fig. 12a).
Immunoblotting
Cells transduced with individual sgRNAs were selected for 2 d in puromycin and cultured for 2 d before collection as described in the base editing screening. Cells treated with siRNAs were subjected to reverse siRNA transfection using firefly (FF) siRNA, BRCA1 siRNA or BRCA2 siRNA at 20 nM and Lipofectamine RNAiMAX (Thermo Fisher Scientific, 13778075) as per the manufacturer’s indications. Cells were trypsinized, washed and resuspended in sample buffer (0.1 M Tris, pH 6.8, 4% SDS, 12% β-mercaptoethanol) at a density of 20,000 cells per microliter. Subsequently, samples were sonicated for 10 s twice and boiled at 95 °C for 5 min before gel electrophoresis. After gel electrophoresis, proteins were transferred onto nitrocellulose membranes. Proteins were detected using the appropriate primary and HRP-conjugated secondary antibodies at a 1:10,000 dilution. Primary antibodies used in this study included mouse-anti-BRCA1 (Santa Cruz Biotechnology, sc-6954, 1:100), rabbit anti-phospho-KAP1 (Bethyl Laboratories, A700-013, 1:1,000), rat anti-tubulin (Novus Biologicals, NB 600-506, 1:50,000) and mouse anti-BRCA2 (Millipore, OP95, 1:1,000).
RNA sequencing
Gamma-irradiated and untreated MCF7-BE3 cells were prepared in parallel to the cells profiled in the DDR364-irradiation screen. Six hours after irradiation, total RNA was extracted with a Quick-RNA Microprep Kit (Zymo Research, R1051), and mRNA was isolated with an NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, E7490L). RNA integrity number (RIN) was quantified with an RNA Pico 6000 assay (Aligent, 5067-1513) on a Bioanalyzer (Aligent 2100, G2939BA). DNA libraries for NGS were prepared with an NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs, E7775) and NEBNext Multiplex Oligos for Illumina (Unique Dual Index UMI Adaptors RNA Set 1) (New England Biolabs, E7416). DNA libraries were quality checked with a DNA 1000 assay (Aligent, 5067-1504) on the Bioanalyzer and then sequenced on a MiSeq platform (Illumina) with a 5% spike-in of PhiX (Azenta Life Sciences, Sequencing-Only). Four replicates were sequenced, and the average transcripts per million (TPM) reads was calculated for the transcripts that we profiled optically with RNAmap.
CRISPRmap and RNAmap primer and padlock probe design
The gene-specific target probes for RNAmap are designed for specificity and minimized off-target binding, conforming to SeqFISH methodologies28,36, using the FISHprobe R package (version 0.4.1; https://github.com/stevexniu/fishprobe). For gene selection and probe extraction, we selected highly expressed gene isoforms from the Human GTEX V8 (ref. 37) and Mouse ENCODE38 tissue expression datasets for probe design. Probe sequences, 20–30 nucleotides in length, were derived from the coding sequence (CDS) and, where necessary, from the untranslated regions (UTRs). We targeted a GC content range of 45–65% or 30–70% for the targeting probes, excluding those with unsuitable GC content or sequences prone to forming homopolymeric runs (such as G-quadruplexes) to maintain optimal hybridization characteristics.
Specificity and off-target minimization: local BLAST39 searches against human and mouse mRNA sequence databases identified probes with off-targets, particularly those with alignments exceeding 10–15 nucleotides with unrelated genes in the transcriptomes and the repetitive DNA using repetitive masks. Tissue-specific expression data from human37 or mouse38 were pivotal in developing a gene copy number table for each tissue type, which informed the exclusion of probes with off-target copy numbers exceeding 15–20 logTPM. For thermal stability and structural integrity, to refine the probe pool by optimizing GC content for enhanced binding affinity, an iterative selection process was employed. Probes were initially ranked in ascending order of their deviation from the target GC content of 55%, starting with the probe exhibiting the greatest deviation. This arrangement continued until no overlapping probes remained. Subsequently, the selection process took into account the calculated melting temperatures (Tm)40. For secondary structure predictions, including pseudoknots, the analysis was conducted under specific conditions: a sodium ion concentration of 0.33 M (equivalent to 2× SSC) and 50% formamide at 37 °C40. Probes with an equilibrium stability lower than 20% were excluded to ensure the formation of stable and specific duplexes. For final probe set selection, the finalized probe set, consisting of 28–32 probes per gene, was optimized to minimize spatial overlap, allowing a maximum of five nucleotides of overlap between adjacent probes. Probes were subjected to stringent filters for equilibrium, and free energy, to refine the probe library. Local BLAST searches within the probe pool identified and mitigated potential cross-hybridization between the selected probes. Genes with insufficient probe numbers were curated manually using a genome browser to guarantee thorough coverage.
CRISPRmap and RNAmap readout probe, CRISPRmap padlock and primer oligo design
For probe generation and off-target screening, starting with a base of 240,000 25mer probes 8, we generated all possible 20mer sequences sequentially. Each of these subsequences was subjected to BLAST screening against human and mouse transcriptomes to exclude any probes with off-target complementarity, and the resulting pool was, thus, reduced to only those probes with zero off-target hits. For optimizing probe performance: to optimize the readout probes’ performance, we calculated their melting temperatures (Tm) and secondary structure predictions39 to refine our selection further, similar to the aforementioned target probes. This ensures that each probe binds to its intended target with high affinity and that the thermal profiles are suitable for our experimental conditions. In scenarios with high mRNA expression, it is vital to prevent overcrowding within any single fluorescence channel. Additionally, based on expression levels in the targeting tissue37,38, we curated the probe sets and their corresponding fluorophores and distributed the signal across multiple channels, promoting distinct visualization of each mRNA molecule. To further minimize the risk of cross-hybridization, we conducted an analysis of readout probe sequences for potential overlaps by performing a local BLAST search against the readout probe pool. This effort led to the identification of 226 20mer DNA sequences, as outlined in Supplementary Table 7, which provides details for each probe, including the 20-nucleotide probe sequence, unique identifier, off-target information, melting temperatures (Tm) and secondary structures. For codebook construction: by employing a Hamming distance approach, similar to the HDM4 code used in MERFISH19, a codebook was constructed with 36 of the aforementioned 226 20mer readout probes. This codebook consists of 319 36-bit codes allocated over 12 hybridization rounds across three channels (488 nm, 561 nm and 640 nm), ensuring that each readout probe would have a unique signature, reducing the possibility of channel crosstalk and fluorescence overlap. This approach aims to enable differentiation of probes even in densely labeled samples, where multiple mRNA molecules are in close proximity. The detailed codebook design is provided in Supplementary Table 7, which includes details such as binary code assignments for each hybridization round and optical channel, indices, a conversion table that relates binary codes to specific probes and sequences linked to each code across channels. For CRISPRmap readout probes, we selected 24 20mers from the aforementioned 226 20mer list. This selected set of 24 probes was split into two sets of 12 probes for the detection of padlocks and primers, respectively. Splint sequences consist of the reverse complement of the primer readout sequences, with an additional universal two bases added at the 5′ end (‘GT’) and the 3′ end (‘AC’) in an attempt to avoid ligation efficiency biases between different splint oligos. Two sets of 54 30mer encoding sequences were generated with similar criteria as the 20mer list and used as padlock or primer encoding sequences. The sequences of these oligos are listed in Supplementary Table 7.
Image processing and analysis
Image storage and stitching
All microscopy images were acquired using Fusion software and saved as IMS files. Each IMS file stores the image as a five-dimensional object in the following order: Resolution, Channel, Z, Y and X. All image montages were stitched using Fusion’s stitching software. For ×60 images, the high-speed setting was used to stitch the image montage and saved as IMS files. For the ×20 images, the high-quality setting using default parameters was used to stitch the image montage and saved as IMS files.
Image rescaling
All images were uploaded to a Google Cloud virtual machine for further image processing and analysis. To read the IMS file and the corresponding metadata, we used the ‘imaris_ims_file_reader’ package. All ×60 montages were analyzed at resolution 3 (1/8 scale of original image), and, for all ×20 images, we used resolution 1 images (1/2 scale of original image). All images were max-projected along the z axis. All max-projected images are three dimensional with the dimensions being Channel, Y and X. The images are in numpy array format and of uint16 data type. Our imaging protocol involves imaging cells at different magnifications based on the resolution of images required. If a particular imaging round was imaged at a different magnification, the images of this round will be of a different size and have a different pixel pitch (pixel-to-micron ratio). To accommodate for this, images were scaled to achieve a consistent pixel pitch using cv2 resize with a bicubic interpolation function. This also ensured that images from all imaging rounds were the same dimensions across X and Y.
Image registration
We register images to a reference image. Across the multiplexed imaging rounds, there are global translational shifts (that is, misalignment of the glass-bottom well plate) as well as local translational shifts (that is, cells slightly shifting between imaging rounds). To finely align the images across all imaging rounds, we calculated the transformation matrices for each round using the TV-L1 (ref. 21) implementation of optical flow on binary nuclei masks derived from DAPI stains. Optical flow calculates Y,X vector shifts across the images for every pixel and performs pixel-level registration. The transformation matrix was then applied to all image channels of that imaging cycle. During registration, all images are converted from uint16 to float64. The images are then converted back to uint16 to reduce the memory usage and speed up image processing. All registered images are three dimensional with the dimensions being Channel, Y and X. Registration quality was estimated using cross-correlation. It is expected that the cross-correlation would decrease with increasing montage size. For our 30 × 30 ×60 montages and 10 × 10 ×20 montages, a cross-correlation of greater than 0.75 was considered good.
Segmentation of cell and nuclear boundaries
To assign detected guides to each cell and quantify nuclear antibody stains, we segmented both the cytoplasmic area and the nuclear area of each cell using Cellpose41. This process was broken into three steps: pre-processing, segmentation and filtering. The EPCAM and DAPI stains were pre-processed by thresholding to maximize the dynamic range of the plasma membrane and nuclear stains in the base editing screening. For other cultured cells, WGA-CF770 (Biotium, 29059) was used for membrane segmentation. For tissue sections, the membrane segmentation was performed on the E-cadherin staining. Typically, this involved setting pixels below the 2nd percentile to 0 and pixels above the 98th percentile to 255.
After pre-processing, images were segmented twice with Cellpose, first to identify cytoplasmic areas and second to identify nuclear areas. The cytoplasmic segmentation run was performed on an image stack containing the EPCAM and DAPI stain and excluded any cytoplasm mask smaller than 5 pixels, whereas the nuclear segmentation was performed only on the DAPI stain with no minimum size requirement. The cell diameter parameter for Cellpose was determined by hand counting and averaging the width and height of 10 randomly sampled cells in pixels. This value was multiplied 1.5× for the cytoplasmic segmentation and 0.75× for the nuclear segmentation.
Once the nuclear and cytoplasmic masks were generated, we filtered out nuclear masks that did not overlap with a cytoplasmic mask and cytoplasmic masks that did not contain a nuclear mask. This ensured that each segmented cytoplasm had one associated segmented nucleus and vice versa. The coordinates for each nuclear and cytoplasmic pair were relabeled with the nuclear ID, which was used as the cell ID from this point onwards. Segmentation quality was validated by both quantifying the percent of proposed cytoplasmic masks retained after filtering for segmented nuclei overlap and visually inspecting the images.
CRISPRmap and RNAmap amplicon detection
To detect amplicons corresponding to CRISPRmap, all registered images corresponding to CRISPRmap readout rounds were processed as follows. Each two-dimensional image (Y and X) underwent contrast stretching to improve the signal-to-noise ratio using the skimage rescale_intensity function. Images were now stored in a list in the order of the readout round and channel (R1-ch1, R1-ch2, R1-ch3, R2-ch1…), with R1 being the first readout round and ch1 being the longest wavelength channel. For each image (readout round and channel combination), spots were identified by using the skimage implementation of the difference of Gaussians method using parameters that maximized the barcode recovery. This implementation outputs an array of coordinates of all spots identified. All the coordinates for spots identified were searched against the cell masks (from cell segmentation), and any spot outside cell masks was discarded from further analysis. Furthermore, if the number of spots within a cell mask was less than 3, then the spots within the cell mask were also discarded from further analysis. This was done to reduce the noise/error in spot detection. All the spots retained for a given round–channel image were stored in an array. This was repeated for each cycle–channel image, and the array of spots retained was stored in a list (with the order being spots for R1-ch1, R1-ch2, R1-ch3, R2-ch1…). Another array was created combining all the retained spots across all imaging round–channel combinations. To eliminate duplicates in the merged array, we used the np.unique function and discarded spots within a radius of 2 pixels. Then, for each spot coordinate in the merged array, we compare the distance of the spot with all the spots detected in a single round–channel combination. If the spot is within a 2-pixel radius, we mark the given round–channel combintion as positive. This was done for all rounds and channels, and, by doing so for each spot, a ‘spot code’ was generated. A spot code essentially maps for a given spot, and round and channel combinations also contain that spot. Once the spot code was generated for all the spots, the spot code for every spot was compared to the predefined barcode designed for every guide. If a spot code matched a barcode, the spot was assigned to the barcode. If a spot did not map to any barcode corresponding to a guide, then the spot was discarded from further analysis. Spot calling was optimized to maximize spots that are assigned to barcodes of the guide library.
Finally, each cell was assigned a barcode based on the spot identity underneath the cell mask, according to the standard CRISPRmap QC as described above. The barcode identity of the cell was stored in a dictionary as well as in the format of an image mask.
Foci and micronuclei detection, cell cycle determination and quantification of antibody stains
For each cell, the sum intensity underneath a given cell mask was calculated for a given antibody stain and stored in a dictionary. The sum intensity underneath the nuclear mask was also calculated and stored in the dictionary. The average intensity of each antibody stain was also calculated by dividing the sum intensity by the total number of pixels underneath the cell/nuclear mask. The raw images then underwent contrast stretching using the rescale_intensity function of skimage. After rescaling the images, foci detection for RAD51, BRCA1, RPA2, γH2AX, 53BP1 and RAD18 was done using the skimage difference of Gaussians method. The total number of foci within the cell mask/nuclear mask was also stored in the dictionary.
To detect and quantify the presence of micronuclei, for every cell mask in the image, the nuclear mask was retrieved. If there was a nuclear mask of less than 100 pixels in area, then the nuclear mask was separately annotated as a small nucleus.
For micronuclei detection, we first performed nuclei segmentation on the DAPI stain using Cellpose. Nuclei masks were generated to define the outline of each nucleus in the image. All the nuclei with a size of less than 100 pixels were marked as ‘micronuclei’. This threshold was determined by manually inspecting the micronuclei captured by the nuclei segmentation. We then subtracted the DAPI signal underneath all nuclei masks by changing the intensity value to 0 for all pixels outlined by the nuclei masks. We removed the background DAPI signal by changing the intensity value to 0 for any pixel with an intensity value lower than 110. To completely remove the residual DAPI signal coming from the DAPI staining of the cell nuclei, we dilated each nuclei mask by 2 pixels using cv2 and then changed the intensity value to 0 for all pixels outlined by the dilated nuclei masks. We finally performed spot calling to identify micronuclei. Based on the coordinates of the spots, the number of spots within a cell mask were identified and included in the dictionary as the number of micronuclei within a cell mask.
Multiplexed immunostaining on cell cycle phase marker proteins was performed to distinguish cells in different cell cycle phases (that is, G0, G1, S/G2 and M phases). Three antibodies were used for cell cycle phase classification—0.5 μg ml−1 rabbit monoclonal anti-Ki-67 (Cell Signaling Technology, 34330), 1 μg ml−1 rabbit monoclonal anti-cyclin A2 (Cell Signaling Technology, 29113SF) and 0.5 μg ml−1 rabbit monoclonal anti-phospho-histone H3 (Cell Signaling Technology, 3475)—and 1 μg ml−1 rabbit monoclonal anti-cyclin B1 (Cell Signaling Technology, 65173SF) was included to further validate the specificity of the cyclin A2 staining. In brief, cell cycle marker signals were quantified for each cell by calculating the average fluorescence intensity under the nuclear mask. The distribution of the average nuclear intensity of each marker was plotted, and a threshold was set to divide cells into positive or negative populations for that marker (shown in Supplementary Fig. 6g). First, cells in the Ki-67− population were classified as G0-phase cells, and cells positive in phospho-histone H3 were classified as M-phase cells; and then cells in the cyclin A2+ population were classified as S/G2-phase cells, and the remaining cells were classified as G1-phase cells.
Statistical analysis on foci features in the base editing screen
For each foci feature that we acquired under each treatment condition in the base editing screen, we calculated a P value based on the Kolmogorov–Smirnov test between the foci count distribution in cells assigned with a given guide identity and all cells assigned with AAVS1-targeting or NTC guide identities (control cells), and then we calculated the L2FC between the average foci count in cells assigned with a given guide identity and the average foci count in all control cells. The Padj was calculated by the Benjamini–Hochberg method. Statistical significance in foci number change is defined by Padj < 0.05 and absolute L2FC > 0.5. Volcano plots were generated to visualize the L2FC and Padj distribution for a given foci feature under a given treatment (Fig. 4g,h,j,k and Supplementary Fig. 8g,h,j,k). We evaluated the foci optical features that we acquired over the ionizing radiation and DNA-damaging agents treatments for each gene variant in the library and counted in how many features a variant resulted in statistically significant change compared to control cells. We tested the number of significant optical features scored by variants in different ClinVar categories with a two-sided Mann–Whitney test (Supplementary Fig. 8m). We also investigated the guides in our library that can lead to the same intended amino acid change. We compared the Pearson correlation of L2FC across all foci features in guides leading to same amino acid changes and guides leading to different amino acid changes with a two-sided independent t-test (Supplementary Fig. 8n).
Sample size analysis for base editing screen hits
In the DDR364 library, we included 72 control (non-targeting or AAVS1-targeting) guides. In the irradiation dataset, we profiled 20,029 control cells over these 72 control guides, whereas the median cell number of a perturbation guide is 281 cells. Therefore, the control population size is roughly 72-fold of a guide on average. In S/G2-phase cells, the median number of cells with perturbation guides is 99 cells. Here, we selected S/G2-phase cells from the top four screen hits of RAD51 foci and BRCA1 foci, respectively, to perform the random sampling analysis. Given the fact that if we vary the number of cells that we image, the size of the control population will still be roughly 72-fold of a given guide, we performed random sampling on the control cells accordingly to maintain this ratio. For each guide, we randomly sampled with replacement for n = 20, 40, …, 200 cells, whereas we sampled n × 72 cells from the control cells in the S/G2 phase, and the P value was calculated by the Kolmogorov–Smirnov test, with statistical significance determined by P < 0.05. The result is shown in Supplementary Fig. 9b,d.
Statistical analysis on co-localized foci features for base editing screen
All co-localization analysis was performed on the irradiated dataset using images at resolution 1 (1/2 scale of the original image), and foci for the six antibody stains (BRCA1, RAD51, γH2AX, 53BP1, RAD18 and RPA2) were detected using the skimage blob_dog method. To account for minor shifts that may not be corrected by registration, foci were considered to co-localize with other foci if the centroids were within a 4-pixel Euclidean distance. This was performed for all 15 (6 choose 2) pairwise combinations of the six antibody stains, and the number of co-localized foci for each pair was then calculated for each single cell that has been assigned with a guide identity (that is, passed QC) by mapping the co-localized foci to its cell nucleus mask.
To determine if the number of co-localized foci observed within the nucleus could be by random overlap of two foci markers, we fixed the coordinates of the first foci marker and permuted the foci for the second marker 10,000 times inside the nucleus. Each time, we calculated the number of co-localized foci using the method described above. The results are shown in Supplementary Fig. 16a. To determine if there was any relevance of cell cycle on the co-localization of foci, we calculated the differences in abundance between a co-localized foci at the S/G2 phase and the G1 phase. The results are shown in Supplementary Fig. 16b. Finally, to calculate the proportion of any given foci co-localizing with another foci, we calculated the proportion of foci co-localized using the formula mean(number of co-localized foci A − B / min(number of co-localized foci A, number of co-localized foci B) per cell). The results are shown in Supplementary Fig. 16c,d.
Void analysis of tissue images
Voids were identified by finding the outermost low-intensity contours where the E-cadherin stain intensity was equal to 113. To set a minimum size for voids, contours with more than 100 boundary pixels were retained as tissue voids. For each retained contour, we gathered the intensity values for the anti-mouse Cd31 stain and the anti-human cleaved PARP1 stain within 20 pixels of the edge of the contour (equidistant inside and outside), which we term ‘boundary stains’. We calculated the 90th percentile of the Cd31 and cPARP boundary stains, and we classified voids as mouse vasculature if the Cd31 value was higher than 113 or as cell death if the cPARP value was higher than 104. Voids negative in both were left unclassified, and one void was classified as double positive.
Clonal purity calculations on tissue images
Following recently reported clonality analysis of barcoded cells27, the clonal score was determined in a cell-centric manner by calculating the local clustering coefficient. In brief, we constructed a 10-nearest neighbors graph for each cell and assigned a P value to each neighborhood by comparing the cell’s same guide clustering coefficient to a table of homotypic clustering coefficients from randomly arranged neighborhoods. This P value was corrected using Bonferroni correction. Cells with Padj < 0.05 were then plotted along with the same guide cells in the 10-nearest neighbor graph to identify clonal regions with significantly higher clustering coefficient.
Bootstrapped Wasserstein distance
We computed a bootstrapped Wasserstein distance to measure the foci expression deviation from perturbation to control guides. We denote \({X}_{\left\{g\right\},{\;j}}\in {Z}^{{|g|}}\) and \({X}_{\left\{c\right\},\;j}\in {Z}^{\,{|c|}}\) as cells undergoing a specific perturbation or control, respectively, where \({|g|}\), \({|c|}\) refer to the corresponding number of cells in each condition. For each perturbation \({g}_{i}\in [\left[G\right]]\) and feature \(j\in\) (RAD51, BRCA1, RPA2, γH2AX, X53BP1 and RAD18), we computed \(W\left({g}_{i},j\right)=\frac{1}{N}\cdot {\Sigma }_{n=1}^{N}{W}_{1}({X}_{\left\{{g}_{i}^{\left(1\right)},\ldots ,{g}_{i}^{\left(S\right)}\right\},j},\,{X}_{\left\{c\right\},{j}})\) as the average 1-Wasserstein distance between guide i and control across \(S=50\) samples with \(N=200\) iterations, where, in each iteration, \(S\), cells \(\left\{{g}_{i}^{\left(1\right)},\ldots ,{g}_{i}^{\left(S\right)}\right\}\subseteq \{{g}_{i}\}\) under guide i are randomly sampled without replacement. As a baseline control, we also computed \(W\left(c,j\right)=\frac{1}{N}\cdot {\Sigma }_{n=1}^{N}{W}_{1}({X}_{\{{c}^{\left(1\right)},\ldots {c}^{\left(S\right)}\},\;j},\,{X}_{\left\{c\right\},\;j})\) as the average 1-Wasserstein distance between randomly subsampled control cells and the full control cell set. The choice of bootstrapping cells is to mitigate the bias introduced by noticeable sample size differences across guides. We report the bootstrapped distances across perturbation guides and the control baseline with violin plots (Supplementary Fig. 3j,k). Highlighted guides were chosen from the aforementioned significant hits with absolute L2FC > 0.5 and Kolmogorov–Smirnov test Padj < 0.05 under Benjamini–Hochberg correction.
Beta-binomial test
We define the data as X ∈ ZN ×2, where Xi:= (ni, ki), where ni denotes the number of cells affected by a given guide, and gi, ki denotes the number of cells exceeding a fixed threshold for a cell-specific continuous or discrete feature. We refer to these cells as ‘positive’ cells. Each guide, gi ∈ [[G]], corresponds to either a control guide or a test guide defined by a mapping φ(gi): [[G]] → {0, 1}, where φ(gi) = 1 corresponds to a test guide. To construct a plausible null hypothesis, we fit a beta-binomial distribution to the control data, Xctrl:= {Xi: φ(gi) = 0}. We use a beta-binomial distribution because we assume the number of positive cells is independently and identically distributed according to a binomial distribution given the number of total cells for a guide. To account for overdispersion attributed to variability between control guides, we place a beta distribution on the success probability of the binomial distribution. For each condition the cells are placed in, we run a separate statistical test because we assume that the rate of positive cells is significantly impacted by the environment, and we would like to test for the significance of specific guides conditioned on the environment.
The model is as follows:
where the parameters α, β are inferred via a maximum likelihood estimation. Then, over the test data, Xtest:= {Xi: φ(gi) = 1}, we compute P values according to a two-tailed test. These P values are then adjusted using the Benjamini–Hochberg correction procedure to control for the FDR. Finally, we reject the null for any corrected P values falling below the significance level of 0.05. To ensure that the null hypothesis is plausible with respect to the control data, we check to see if the computed P values are uniformly distributed over the [0, 1] interval. The proportions, total number of cells and adjusted P values for each test guide are reported in Supplementary Table 7.
Statistics and reproducibility
Histograms were plotted with the histplot function, boxplots with the boxplot function, scatter plots and volcano plots with the scatterplot function, ECDF plots with the ecdfplot function and violin plots with the violinplot function in the seaborn package (0.11.1) in Python. Two-sided Mann–Whitney U-tests and Studentʼs t-tests were performed to test the difference between the distributions using the statannotations package in Python. *P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.0001. The difference in foci number distribution was tested by a two-sided Kolmogorov–Smirnov test using the ks.test function in R, and the P values were adjusted by the Benjamini–Hochberg method. *Padj < 0.05, **Padj < 0.01, ***Padj < 0.001, ****Padj < 0.0001. Guides with Padj < 0.05 and an absolute L2FC > 0.5 were regarded as statistically significant. Fisherʼs exact test for gene enrichment was performed with the fisher.test function in R, and the P values were adjusted by the Benjamini–Hochberg method. Genes with Padj < 0.05 were regarded as statistically significant. Heatmaps for optical feature correlations and hierarchical clustering of guides were generated with the clustermap function in the seaborn package (0.11.1) in Python, and the method ‘complete’ was used for clustering. Schematics were generated in BioRender. All representative images shown in this paper were repeated in at least three technical replicates with similar results, unless otherwise specified in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All raw imaging data for the CRISPRmap screens mentioned in this paper were deposited into the BioImage Archive database under accession number S-BIAD985 and are available at the following URL: https://www.ebi.ac.uk/biostudies/BioImages/studies/S-BIAD985 (ref. 42). The ClinVar database for variant characterization is available at https://www.ncbi.nlm.nih.gov/clinvar/.
Code availability
CRISPRmap analysis was performed using customized code, which is available at the following URL: https://github.com/GaublommeLab/CRISPRmap-Pipeline (ref. 43).
Change history
05 March 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41587-025-02594-z
References
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866 (2016).
Feldman, D. et al. Pooled genetic perturbation screens with image-based phenotypes. Nat. Protoc. 17, 476–512 (2022).
Feldman, D. et al. Optical pooled screens in human cells. Cell 179, 787–799 (2019).
Funk, L. et al. The phenotypic landscape of essential human genes. Cell 185, 4634–4653 (2022).
Wang, C., Lu, T., Emanuel, G., Babcock, H. P. & Zhuang, X. Imaging-based pooled CRISPR screening reveals regulators of lncRNA localization. Proc. Natl Acad. Sci. USA 116, 10842–10851 (2019).
Cuella-Martin, R. et al. Functional interrogation of DNA damage response variants with base editing screens. Cell 184, 1081–1097 (2021).
Hanna, R. E. et al. Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080 (2021).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Matejcic, M. et al. Pathogenic variants in cancer predisposition genes and prostate cancer risk in men of African ancestry. JCO Precis. Oncol. 4, 32–43 (2020).
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Liu, L. F. et al. Mechanism of action of camptothecin. Ann. N. Y. Acad. Sci. 922, 1–10 (2000).
Goulooze, S. C., Cohen, A. F. & Rissmann, R. Olaparib. Br. J. Clin. Pharmacol. 81, 171–173 (2016).
Dasari, S. & Tchounwou, P. B. Cisplatin in cancer therapy: molecular mechanisms of action. Eur. J. Pharmacol. 740, 364–378 (2014).
Montecucco, A., Zanetta, F. & Biamonti, G. Molecular mechanisms of etoposide. EXCLI J. 14, 95–108 (2015).
Dhainaut, M. et al. Spatial CRISPR genomics identifies regulators of the tumor microenvironment. Cell 185, 1223–1239 (2022).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Shah, S. et al. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174, 363–376 (2018).
Wedel, A., Pock, T., Zach, C., Bischof, H. & Cremers, D. An improved algorithm for TV-L1 optical flow. In Statistical and Geometrical Approaches to Visual Motion Analysis. Lecture Notes in Computer Science (eds Cremers, D., Rosenhahn, B., Yuille, A. L. & Schmidt, F. R.). https://doi.org/10.1007/978-3-642-03061-1_2 (Springer, 2009).
Radtke, A. J. et al. IBEX: a versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc. Natl Acad. Sci. USA 117, 33455–33465 (2020).
McManus, K. J. & Hendzel, M. J. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol. Biol. Cell 16, 5013–5025 (2005).
Landrum, M. J. et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 48, D835–D844 (2020).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Kweon, J. et al. A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene 39, 30–35 (2020).
Rovira-Clave, X. et al. Spatial epitope barcoding reveals clonal tumor patch behaviors. Cancer Cell 40, 1423–1439 (2022).
Eng, C. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019).
Carlson, R. J., Leiken, M. D., Guna, A., Hacohen, N. & Blainey, P. C. A genome-wide optical pooled screen reveals regulators of cellular antiviral responses. Proc. Natl Acad. Sci. USA 120, e2210623120 (2023).
Askary, A. et al. In situ readout of DNA barcodes and single base edits facilitated by in vitro transcription. Nat. Biotechnol. 38, 66–75 (2020).
Pantazis, C. B. et al. A reference human induced pluripotent stem cell line for large-scale collaborative studies. Cell Stem Cell 29, 1685–1702 (2022).
Bos, P. H. et al. Development of MAP4 kinase inhibitors as motor neuron-protecting agents. Cell Chem. Biol. 26, 1703–1715 (2019).
Maury, Y. et al. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 33, 89–96 (2015).
CRISPick. https://portals.broadinstitute.org/gppx/crispick/public
Omelina, E. S., Ivankin, A. V., Letiagina, A. E. & Pindyurin, A. V. Optimized PCR conditions minimizing the formation of chimeric DNA molecules from MPRA plasmid libraries. BMC Genomics 20, 536 (2019).
Eng, C. L., Shah, S., Thomassie, J. & Cai, L. Profiling the transcriptome with RNA SPOTs. Nat. Methods 14, 1153–1155 (2017).
Consortium, G. T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Mouse ENCODE Consortiumet al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 13, 418 (2012).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Zadeh, J. N. et al. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Gu, J. et al. Mapping multi-modal phenotypes to perturbations in cells and tissue with CRISPRmap. https://www.ebi.ac.uk/biostudies/BioImages/studies/S-BIAD985?query=S-BIAD985 (2024).
Gu, J. et al. Mapping multi-modal phenotypes to perturbations in cells and tissue with CRISPRmap. https://github.com/GaublommeLab/CRISPRmap-Pipeline (2024).
Acknowledgements
We would like to thank N. Barolini for figure design. We acknowledge support by National Institutes of Health (NIH), National Cancer Institute grants 1DP2CA281605 to J.T.G. and R21HG012639 to E.A.; NIH National Human Genome Research Institute grant R01HG012875 to E.A.; the Cancer Research Institute Lloyd J. Old STAR award; R33 CA267219 and UG3 NS132139 to D.A.L.; and NIH grants R01CA197774 and R01CA227450 to A.C.
Author information
Authors and Affiliations
Contributions
J.T.G. and J.G. conceived the GFP knockout pilot study. J.T.G., A.C. and J.G. conceived the base editing study. J.T.G., E.M.C. and J.G. conceived the tissue profiling study. J.T.G. conceived CRISPRmap and RNAmap, and J.G. implemented CRISPRmap. J.C., A.D., R.G. and C.C. contributed to experiments that informed the development of RNAmap, under the supervision of J.T.G. J.G., Y.S., G.L. and A.T. performed experiments, under the supervision of A.C. and J.T.G. S.H., K.J. and L.P.-B. performed mouse work, under the supervision of E.M.C. J.A.C. and J.G. performed the iPSC and iMN work. M.G.P. and J.G. performed the hESC work, under the supervision of J.T.G. and H.S. A.I., B.W., J.G., N.K. and M.P. developed the image analysis pipeline, under the supervision of J.T.G. X.N. developed the FISHprobe pipeline, under the supervision of D.L. and J.T.G. J.G., A.I., Y.J., J.Y.Z. and J.H. performed analysis, under the supervision of E.A., A.C. and J.T.G. J.G., B.W., N.K., Y.J. and J.H. generated figures, under the supervision of E.A., A.C. and J.T.G. J.G. and J.T.G. wrote the manuscript, with input from all authors.
Corresponding author
Ethics declarations
Competing interests
Columbia University has filed a patent application related to this work, for which J.T.G. is an inventor. E.M.C.ʼs laboratory receives support from Novartis. D.A.L. has served as a consultant for AbbVie, AstraZeneca and Illumina and is on the scientific advisory boards of Mission Bio, Pangea, Alethiomics and C2i Genomics. D.A.L. has also received prior research funding from Bristol Myers Squibb, 10x Genomics, Ultima Genomics and Illumina, all unrelated to the current study. The remaining authors declare no competing interests.
Ethics
All human iPSCs and embyonic stem cells (ESCs) used in this study were previously generated and reported to be derived from material obtained under informed consent and appropriate ethical approvals. The human ESC (RUES2) is approved by the National Institutes of Health (NIH) Intramural Embryonic Stem Cell Research Oversight Committee. Animal care and experimental procedures were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Columbia University Institutional Animal Care and Use Committee (protocol AC-AABT8654).
Peer review
Peer review information
Nature Biotechnology thanks Brian Brown, Howard Chang and Prisca Liberali for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Information
Supplementary Figs. 1–17, supplementary Source Data files and supplementary table information.
Supplementary Table 1
CRISPRmap cost in DDR364 screening.
Supplementary Table 2
GFP-pilot analysis.
Supplementary Table 3
Base editing screen analysis.
Supplementary Table 4
OE19 tissue analysis.
Supplementary Table 5
GFP-pilot library design.
Supplementary Table 6
DDR364 library design.
Supplementary Table 7
Oligonucleotide sequences in CRISPRmap and RNAmap.
Supplementary Table 8
Protein and RNA quantification of irradiated cells in DDR364 screen.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gu, J., Iyer, A., Wesley, B. et al. Mapping multimodal phenotypes to perturbations in cells and tissue with CRISPRmap. Nat Biotechnol 43, 1101–1115 (2025). https://doi.org/10.1038/s41587-024-02386-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41587-024-02386-x
This article is cited by
-
Cellular neighborhoods in cancer
Nature Cancer (2026)
-
Interpretation, extrapolation and perturbation of single cells
Nature Reviews Genetics (2026)
-
Bridging the variant-to-function gap in type 2 diabetes: advances and challenges
Diabetologia (2026)
-
CRISPR-Based Functional Genomics in Pluripotent Stem Cells
Stem Cell Reviews and Reports (2026)
-
Integrated in vivo combinatorial functional genomics and spatial transcriptomics of tumours to decode genotype-to-phenotype relationships
Nature Biomedical Engineering (2025)